New insights into the interplay between the translation machinery and nonsense-mediated mRNA decay factors
- PMID: 29626148
- PMCID: PMC6008592
- DOI: 10.1042/BST20170427
New insights into the interplay between the translation machinery and nonsense-mediated mRNA decay factors
Abstract
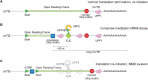
Faulty mRNAs with a premature stop codon (PTC) are recognized and degraded by nonsense-mediated mRNA decay (NMD). Recognition of a nonsense mRNA depends on translation and on the presence of NMD-enhancing or the absence of NMD-inhibiting factors in the 3'-untranslated region. Our review summarizes our current understanding of the molecular function of the conserved NMD factors UPF3B and UPF1, and of the anti-NMD factor Poly(A)-binding protein, and their interactions with ribosomes translating PTC-containing mRNAs. Our recent discovery that UPF3B interferes with human translation termination and enhances ribosome dissociation in vitro, whereas UPF1 is inactive in these assays, suggests a re-interpretation of previous experiments and modification of prevalent NMD models. Moreover, we discuss recent work suggesting new functions of the key NMD factor UPF1 in ribosome recycling, inhibition of translation re-initiation and nascent chain ubiquitylation. These new findings suggest that the interplay of UPF proteins with the translation machinery is more intricate than previously appreciated, and that this interplay quality-controls the efficiency of termination, ribosome recycling and translation re-initiation.
Keywords: UPF proteins; mRNA quality control; nonsense-mediated mRNA decay; ribosome; translation termination.
© 2018 The Author(s).
Conflict of interest statement
The Authors declare that there are no competing interests associated with the manuscript.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

