Specific and Sensitive Primers Developed by Comparative Genomics to Detect Bacterial Pathogens in Grains
- PMID: 29628816
- PMCID: PMC5880354
- DOI: 10.5423/PPJ.OA.11.2017.0250
Specific and Sensitive Primers Developed by Comparative Genomics to Detect Bacterial Pathogens in Grains
Abstract
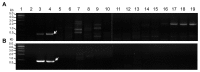
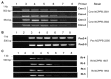
Accurate and rapid detection of bacterial plant pathogen is the first step toward disease management and prevention of pathogen spread. Bacterial plant pathogens Clavibacter michiganensis subsp. nebraskensis (Cmn), Pantoea stewartii subsp. stewartii (Pss), and Rathayibacter tritici (Rt) cause Goss's bacterial wilt and blight of maize, Stewart's wilt of maize and spike blight of wheat and barley, respectively. The bacterial diseases are not globally distributed and not present in Korea. This study adopted comparative genomics approach and aimed to develop specific primer pairs to detect these three bacterial pathogens. Genome comparison among target pathogens and their closely related bacterial species generated 15-20 candidate primer pairs per bacterial pathogen. The primer pairs were assessed by a conventional PCR for specificity against 33 species of Clavibacter, Pantoea, Rathayibacter, Pectobacterium, Curtobacterium. The investigation for specificity and sensitivity of the primer pairs allowed final selection of one or two primer pairs per bacterial pathogens. In our assay condition, a detection limit of Pss and Cmn was 2 pg/μl of genomic DNA per PCR reaction, while the detection limit for Rt primers was higher. The selected primers could also detect bacterial cells up to 8.8 × 103 cfu to 7.84 × 104 cfu per gram of grain seeds artificially infected with corresponding bacterial pathogens. The primer pairs and PCR assay developed in this study provide an accurate and rapid detection method for three bacterial pathogens of grains, which can be used to investigate bacteria contamination in grain seeds and to ultimately prevent pathogen dissemination over countries.
Keywords: Clavibacter michiganensis subsp. nebraskensis; PCR detection; Pantoea stewartii subsp. stewartii; Rathayibacter tritici; comparative genomics.
Figures



References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

