A Statistically Representative Atlas for Mapping Neuronal Circuits in the Drosophila Adult Brain
- PMID: 29628885
- PMCID: PMC5876320
- DOI: 10.3389/fninf.2018.00013
A Statistically Representative Atlas for Mapping Neuronal Circuits in the Drosophila Adult Brain
Abstract
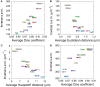
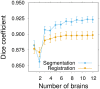
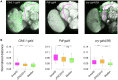
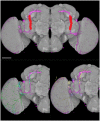
Imaging the expression patterns of reporter constructs is a powerful tool to dissect the neuronal circuits of perception and behavior in the adult brain of Drosophila, one of the major models for studying brain functions. To date, several Drosophila brain templates and digital atlases have been built to automatically analyze and compare collections of expression pattern images. However, there has been no systematic comparison of performances between alternative atlasing strategies and registration algorithms. Here, we objectively evaluated the performance of different strategies for building adult Drosophila brain templates and atlases. In addition, we used state-of-the-art registration algorithms to generate a new group-wise inter-sex atlas. Our results highlight the benefit of statistical atlases over individual ones and show that the newly proposed inter-sex atlas outperformed existing solutions for automated registration and annotation of expression patterns. Over 3,000 images from the Janelia Farm FlyLight collection were registered using the proposed strategy. These registered expression patterns can be searched and compared with a new version of the BrainBaseWeb system and BrainGazer software. We illustrate the validity of our methodology and brain atlas with registration-based predictions of expression patterns in a subset of clock neurons. The described registration framework should benefit to brain studies in Drosophila and other insect species.
Keywords: Drosophila adult brain; anatomical atlas; atlas-based image segmentation; average brain template; brain mapping; confocal microscopy; diffeomorphic image registration.
Figures






References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

