FANCM Limits Meiotic Crossovers in Brassica Crops
- PMID: 29628933
- PMCID: PMC5876677
- DOI: 10.3389/fpls.2018.00368
FANCM Limits Meiotic Crossovers in Brassica Crops
Erratum in
-
Corrigendum: FANCM Limits Meiotic Crossovers in Brassica Crops.Front Plant Sci. 2020 Dec 4;11:604728. doi: 10.3389/fpls.2020.604728. eCollection 2020. Front Plant Sci. 2020. PMID: 33343604 Free PMC article.
Abstract
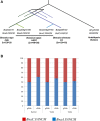
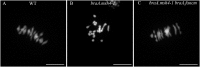
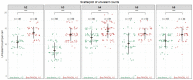
Meiotic crossovers (COs) are essential for proper chromosome segregation and the reshuffling of alleles during meiosis. In WT plants, the number of COs is usually small, which limits the genetic variation that can be captured by plant breeding programs. Part of this limitation is imposed by proteins like FANCM, the inactivation of which results in a 3-fold increase in COs in Arabidopsis thaliana. Whether the same holds true in crops needed to be established. In this study, we identified EMS induced mutations in FANCM in two species of economic relevance within the genus Brassica. We showed that CO frequencies were increased in fancm mutants in both diploid and tetraploid Brassicas, Brassica rapa and Brassica napus respectively. In B. rapa, we observed a 3-fold increase in the number of COs, equal to the increase observed previously in Arabidopsis. In B. napus we observed a lesser but consistent increase (1.3-fold) in both euploid (AACC) and allohaploid (AC) plants. Complementation tests in A. thaliana suggest that the smaller increase in crossover frequency observed in B. napus reflects residual activity of the mutant C copy of FANCM. Altogether our results indicate that the anti-CO activity of FANCM is conserved across the Brassica, opening new avenues to make a wider range of genetic diversity accessible to crop improvement.
Keywords: Brassica; FANCM; TILLING; Translational biology; meiotic crossover; plant breeding; polyploidy.
Figures

References
-
- Anderson L. K., Lohmiller L. D., Tang X., Hammond D. B., Javernick L., Shearer L., et al. (2014). Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proc. Natl. Acad. Sci. U.S.A. 111, 13415–13420. 10.1073/pnas.1406846111 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

