A Method for Targeted 16S Sequencing of Human Milk Samples
- PMID: 29630048
- PMCID: PMC5933250
- DOI: 10.3791/56974
A Method for Targeted 16S Sequencing of Human Milk Samples
Abstract
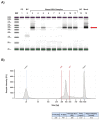
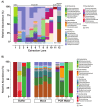
Studies of microbial communities have become widespread with the development of relatively inexpensive, rapid, and high throughput sequencing. However, as with all these technologies, reproducible results depend on a laboratory workflow that incorporates appropriate precautions and controls. This is particularly important with low-biomass samples where contaminating bacterial DNA can generate misleading results. This article details a semi-automated workflow to identify microbes from human breast milk samples using targeted sequencing of the 16S ribosomal RNA (rRNA) V4 region on a low- to mid-throughput scale. The protocol describes sample preparation from whole milk including: sample lysis, nucleic acid extraction, amplification of the V4 region of the 16S rRNA gene, and library preparation with quality control measures. Importantly, the protocol and discussion consider issues that are salient to the preparation and analysis of low-biomass samples including appropriate positive and negative controls, PCR inhibitor removal, sample contamination by environmental, reagent, or experimental sources, and experimental best practices designed to ensure reproducibility. While the protocol as described is specific to human milk samples, it is adaptable to numerous low- and high-biomass sample types, including samples collected on swabs, frozen neat, or stabilized in a preservation buffer.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources