ERASE-Seq: Leveraging replicate measurements to enhance ultralow frequency variant detection in NGS data
- PMID: 29630678
- PMCID: PMC5890993
- DOI: 10.1371/journal.pone.0195272
ERASE-Seq: Leveraging replicate measurements to enhance ultralow frequency variant detection in NGS data
Abstract
The accurate detection of ultralow allele frequency variants in DNA samples is of interest in both research and medical settings, particularly in liquid biopsies where cancer mutational status is monitored from circulating DNA. Next-generation sequencing (NGS) technologies employing molecular barcoding have shown promise but significant sensitivity and specificity improvements are still needed to detect mutations in a majority of patients before the metastatic stage. To address this we present analytical validation data for ERASE-Seq (Elimination of Recurrent Artifacts and Stochastic Errors), a method for accurate and sensitive detection of ultralow frequency DNA variants in NGS data. ERASE-Seq differs from previous methods by creating a robust statistical framework to utilize technical replicates in conjunction with background error modeling, providing a 10 to 100-fold reduction in false positive rates compared to published molecular barcoding methods. ERASE-Seq was tested using spiked human DNA mixtures with clinically realistic DNA input quantities to detect SNVs and indels between 0.05% and 1% allele frequency, the range commonly found in liquid biopsy samples. Variants were detected with greater than 90% sensitivity and a false positive rate below 0.1 calls per 10,000 possible variants. The approach represents a significant performance improvement compared to molecular barcoding methods and does not require changing molecular reagents.
Conflict of interest statement
Figures

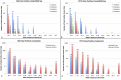
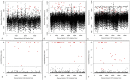
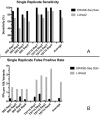
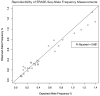
References
-
- Rehm HL. Disease-targeted sequencing: a cornerstone in the clinic. Nat Rev Genet. 2013;14(4):295–300. doi: 10.1038/nrg3463 - DOI - PMC - PubMed
-
- Boycott KM, Vanstone MR, Bulman DE, MacKenzie AE. Rare-disease genetics in the era of next-generation sequencing: discovery to translation. Nat Rev Genet. 2013;14(10):681–91. doi: 10.1038/nrg3555 . - DOI - PubMed
-
- Stadler ZK, Schrader KA, Vijai J, Robson ME, Offit K. Cancer genomics and inherited risk. J Clin Oncol. 2014;32(7):687–98. doi: 10.1200/JCO.2013.49.7271 . - DOI - PMC - PubMed
-
- Perera MA, Gamazon E, Cavallari LH, Patel SR, Poindexter S, Kittles RA, et al. The missing association: sequencing-based discovery of novel SNPs in VKORC1 and CYP2C9 that affect warfarin dose in African Americans. Clin Pharmacol Ther. 2011;89(3):408–15. doi: 10.1038/clpt.2010.322 - DOI - PMC - PubMed
-
- Frampton GM, Fichtenholtz A, Otto GA, Wang K, Downing SR, He J, et al. Development and validation of a clinical cancer genomic profiling test based on massively parallel DNA sequencing. Nat Biotechnol. 2013;31(11):1023–31. doi: 10.1038/nbt.2696 . - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

