Caloric restriction mitigates age-associated hippocampal differential CG and non-CG methylation
- PMID: 29631215
- PMCID: PMC5955819
- DOI: 10.1016/j.neurobiolaging.2018.03.009
Caloric restriction mitigates age-associated hippocampal differential CG and non-CG methylation
Abstract
Brain aging is marked by cognitive decline and susceptibility to neurodegeneration. Calorie restriction (CR) increases neurogenesis, improves memory function, and protects from age-associated neurological disorders. Epigenetic mechanisms, including DNA methylation, are vital to normal central nervous system cellular and memory functions and are dysregulated with aging. The beneficial effects of CR have been proposed to work through epigenetic processes, but this is largely unexplored. We therefore tested whether life long CR prevents age-related hippocampal DNA methylation changes. Hippocampal DNA from young (3 months) and old (24 months) male mice fed ad libitum and 24-month-old mice fed a 40% calorie-restricted diet from 3 months of age were examined by genome-wide bisulfite sequencing to measure methylation with base specificity. Over 27 million CG and CH (non-CG) sites were examined. Of the ∼40,000 differentially methylated CG and ∼80,000 CH sites with aging, >1/3 were prevented by CR and were found across genomic regulatory regions and gene pathways. CR also caused alterations to CG and CH methylation at sites not differentially methylated with aging, and these CR-specific changes demonstrated a different pattern of regulatory element and gene pathway enrichment than those affected by aging. CR-specific DNA methyltransferase 1 and Tet methylcytosine dioxygenase 3 promoter hypermethylation corresponded to reduced gene expression. These findings demonstrate that CR attenuates age-related CG and CH hippocampal methylation changes, in combination with CR-specific methylation that may also contribute to the neuroprotective effects of CR. The prevention of age-related methylation alterations is also consistent with the prolongevity effects of CR working through an epigenetic mechanism.
Keywords: Aging; Caloric restriction; DNA methylation; Epigenetics; Hippocampus.
Copyright © 2018 Elsevier Inc. All rights reserved.
Conflict of interest statement
Figures

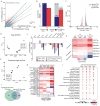


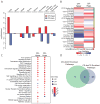
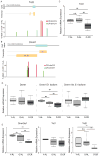
References
-
- Cedar H, Bergman Y. Linking DNA methylation and histone modification: patterns and paradigms. Nature reviews Genetics. 2009;10(5):295–304. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

