PARP-1 protects against colorectal tumor induction, but promotes inflammation-driven colorectal tumor progression
- PMID: 29632181
- PMCID: PMC5924876
- DOI: 10.1073/pnas.1712345115
PARP-1 protects against colorectal tumor induction, but promotes inflammation-driven colorectal tumor progression
Abstract
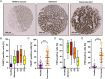
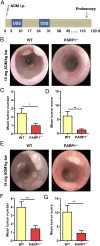
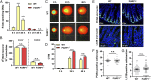
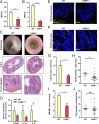
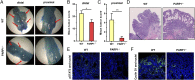
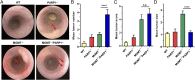
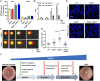
Colorectal cancer (CRC) is one of the most common tumor entities, which is causally linked to DNA repair defects and inflammatory bowel disease (IBD). Here, we studied the role of the DNA repair protein poly(ADP-ribose) polymerase-1 (PARP-1) in CRC. Tissue microarray analysis revealed PARP-1 overexpression in human CRC, correlating with disease progression. To elucidate its function in CRC, PARP-1 deficient (PARP-1-/-) and wild-type animals (WT) were subjected to azoxymethane (AOM)/ dextran sodium sulfate (DSS)-induced colorectal carcinogenesis. Miniendoscopy showed significantly more tumors in WT than in PARP-1-/- mice. Although the lack of PARP-1 moderately increased DNA damage, both genotypes exhibited comparable levels of AOM-induced autophagy and cell death. Interestingly, miniendoscopy revealed a higher AOM/DSS-triggered intestinal inflammation in WT animals, which was associated with increased levels of innate immune cells and proinflammatory cytokines. Tumors in WT animals were more aggressive, showing higher levels of STAT3 activation and cyclin D1 up-regulation. PARP-1-/- animals were then crossed with O6-methylguanine-DNA methyltransferase (MGMT)-deficient animals hypersensitive to AOM. Intriguingly, PARP-1-/-/MGMT-/- double knockout (DKO) mice developed more, but much smaller tumors than MGMT-/- animals. In contrast to MGMT-deficient mice, DKO animals showed strongly reduced AOM-dependent colonic cell death despite similar O6-methylguanine levels. Studies with PARP-1-/- cells provided evidence for increased alkylation-induced DNA strand break formation when MGMT was inhibited, suggesting a role of PARP-1 in the response to O6-methylguanine adducts. Our findings reveal PARP-1 as a double-edged sword in colorectal carcinogenesis, which suppresses tumor initiation following DNA alkylation in a MGMT-dependent manner, but promotes inflammation-driven tumor progression.
Keywords: DNA repair; PARP-1; colorectal carcinogenesis; intestinal inflammation; mouse models.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Fahrer J, Kaina B. O6-methylguanine-DNA methyltransferase in the defense against N-nitroso compounds and colorectal cancer. Carcinogenesis. 2013;34:2435–2442. - PubMed
-
- Fahrer J, et al. DNA repair by MGMT, but not AAG, causes a threshold in alkylation-induced colorectal carcinogenesis. Carcinogenesis. 2015;36:1235–1244. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

