Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector
- PMID: 29632371
- PMCID: PMC5948154
- DOI: 10.1038/s41591-018-0004-z
Gain of toxic apolipoprotein E4 effects in human iPSC-derived neurons is ameliorated by a small-molecule structure corrector
Abstract
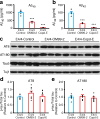
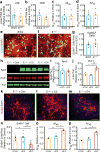
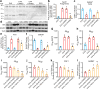
Efforts to develop drugs for Alzheimer's disease (AD) have shown promise in animal studies, only to fail in human trials, suggesting a pressing need to study AD in human model systems. Using human neurons derived from induced pluripotent stem cells that expressed apolipoprotein E4 (ApoE4), a variant of the APOE gene product and the major genetic risk factor for AD, we demonstrated that ApoE4-expressing neurons had higher levels of tau phosphorylation, unrelated to their increased production of amyloid-β (Aβ) peptides, and that they displayed GABAergic neuron degeneration. ApoE4 increased Aβ production in human, but not in mouse, neurons. Converting ApoE4 to ApoE3 by gene editing rescued these phenotypes, indicating the specific effects of ApoE4. Neurons that lacked APOE behaved similarly to those expressing ApoE3, and the introduction of ApoE4 expression recapitulated the pathological phenotypes, suggesting a gain of toxic effects from ApoE4. Treatment of ApoE4-expressing neurons with a small-molecule structure corrector ameliorated the detrimental effects, thus showing that correcting the pathogenic conformation of ApoE4 is a viable therapeutic approach for ApoE4-related AD.
Figures



References
-
- Corder E, et al. Gene dose of apolipoprotein E type 4 allele and the risk of Alzheimer’s disease in late onset families. Science. 1993;261:921–923. - PubMed
-
- Saunders AM, et al. Association of apolipoprotein E allele e4 with late-onset familial and sporadic Alzheimer’s disease. Neurology. 1993;43:1467–1472. - PubMed
-
- Farrer LA, et al. Effects of age, sex, and ethnicity on the association between apolipoprotein E genotype and Alzheimer disease. A meta-analysis. J. Am. Med. Assoc. 1997;278:1349–1356. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

