Moving Beyond ERP Components: A Selective Review of Approaches to Integrate EEG and Behavior
- PMID: 29632480
- PMCID: PMC5879117
- DOI: 10.3389/fnhum.2018.00106
Moving Beyond ERP Components: A Selective Review of Approaches to Integrate EEG and Behavior
Abstract
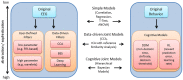
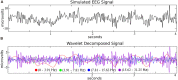
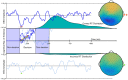
Relationships between neuroimaging measures and behavior provide important clues about brain function and cognition in healthy and clinical populations. While electroencephalography (EEG) provides a portable, low cost measure of brain dynamics, it has been somewhat underrepresented in the emerging field of model-based inference. We seek to address this gap in this article by highlighting the utility of linking EEG and behavior, with an emphasis on approaches for EEG analysis that move beyond focusing on peaks or "components" derived from averaging EEG responses across trials and subjects (generating the event-related potential, ERP). First, we review methods for deriving features from EEG in order to enhance the signal within single-trials. These methods include filtering based on user-defined features (i.e., frequency decomposition, time-frequency decomposition), filtering based on data-driven properties (i.e., blind source separation, BSS), and generating more abstract representations of data (e.g., using deep learning). We then review cognitive models which extract latent variables from experimental tasks, including the drift diffusion model (DDM) and reinforcement learning (RL) approaches. Next, we discuss ways to access associations among these measures, including statistical models, data-driven joint models and cognitive joint modeling using hierarchical Bayesian models (HBMs). We think that these methodological tools are likely to contribute to theoretical advancements, and will help inform our understandings of brain dynamics that contribute to moment-to-moment cognitive function.
Keywords: EEG; ERP; blind source separation; canonical correlations analysis; deep learning; hierarchical Bayesian model; partial least squares; representational similarity analysis.
Figures




References
-
- Andersson C. A., Bro R. (2000). The N-way toolbox for MATLAB. Chemom. Intell. Lab. Syst. 52, 1–4. 10.1016/S0169-7439(00)00071-X - DOI
-
- Basar E. (1999). Brain Function and Oscillations, Principles and Approaches, 1. Berlin: Springer.
-
- Bashivan P., Rish I., Yeasin M., Codella N. (2016). “Learning representations from EEG with deep recurrent-convolutional neural networks,” in Proceedings of the International Conference on Learning Representations (San Juan), 2–4.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

