Sleep loss disrupts Arc expression in dentate gyrus neurons
- PMID: 29635031
- PMCID: PMC6174112
- DOI: 10.1016/j.nlm.2018.04.006
Sleep loss disrupts Arc expression in dentate gyrus neurons
Abstract
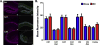
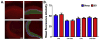
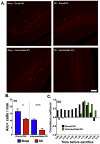
Sleep loss affects many aspects of cognition, and memory consolidation processes occurring in the hippocampus seem particularly vulnerable to sleep loss. The immediate-early gene Arc plays an essential role in both synaptic plasticity and memory formation, and its expression is altered by sleep. Here, using a variety of techniques, we have characterized the effects of brief (3-h) periods of sleep vs. sleep deprivation (SD) on the expression of Arc mRNA and Arc protein in the mouse hippocampus and cortex. By comparing the relative abundance of mature Arc mRNA with unspliced pre-mRNA, we see evidence that during SD, increases in Arc across the cortex, but not hippocampus, reflect de novo transcription. Arc increases in the hippocampus during SD are not accompanied by changes in pre-mRNA levels, suggesting that increases in mRNA stability, not transcription, drives this change. Using in situ hybridization (together with behavioral observation to quantify sleep amounts), we find that in the dorsal hippocampus, SD minimally affects Arc mRNA expression, and decreases the number of dentate gyrus (DG) granule cells expressing Arc. This is in contrast to neighboring cortical areas, which show large increases in neuronal Arc expression after SD. Using immunohistochemistry, we find that Arc protein expression is also differentially affected in the cortex and DG with SD - while larger numbers of cortical neurons are Arc+, fewer DG granule cells are Arc+, relative to the same regions in sleeping mice. These data suggest that with regard to expression of plasticity-regulating genes, sleep (and SD) can have differential effects in hippocampal and cortical areas. This may provide a clue regarding the susceptibility of performance on hippocampus-dependent tasks to deficits following even brief periods of sleep loss.
Keywords: In situ hybridization; Sleep deprivation; Synaptic plasticity; Transcription; Translation; pre-mRNA.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

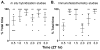



References
-
- Acosta-Pena E, Camacho-Abrego I, Megarejo-Gutierrez M, Flores G, Drucker-Colin R, Garcia-Garcia F. Sleep deprivation induces differential morphological changes in the hippocampus and prefrontal cortex in young and old rats. Synapse. 2015;69:15–25. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

