New Methods for Inferring the Distribution of Fitness Effects for INDELs and SNPs
- PMID: 29635416
- PMCID: PMC5967470
- DOI: 10.1093/molbev/msy054
New Methods for Inferring the Distribution of Fitness Effects for INDELs and SNPs
Abstract
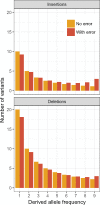
Small insertions and deletions (INDELs; ≤50 bp) are the most common type of variability after single nucleotide polymorphism (SNP). However, compared with SNPs, we know little about the distribution of fitness effects (DFE) of new INDEL mutations and how prevalent adaptive INDEL substitutions are. Studying INDELs has been difficult partly because identifying ancestral states at these sites is error-prone and misidentification can lead to severely biased estimates of the strength of selection. To solve these problems, we develop new maximum likelihood methods, which use polymorphism data to simultaneously estimate the DFE, the mutation rate, and the misidentification rate. These methods are applicable to both INDELs and SNPs. Simulations show that they can provide highly accurate results. We applied the methods to an INDEL polymorphism data set in Drosophila melanogaster. We found that the DFE for polymorphic INDELs in protein-coding regions is bimodal, with the variants being either nearly neutral or strongly deleterious. Based on the DFE, we estimated that 71.5-83.7% of the INDEL substitutions that took place along the D. melanogaster lineage were fixed by positive selection, which is comparable with the prevalence of adaptive substitutions at nonsynonymous sites. The new methods have been implemented in the software package anavar.
Figures
References
-
- Andolfatto P. 2005. Adaptive evolution of non-coding DNA in Drosophila. Nature 437(7062):1149–1152. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

