Metabolomic studies identify changes in transmethylation and polyamine metabolism in a brain-specific mouse model of tuberous sclerosis complex
- PMID: 29635516
- PMCID: PMC5985733
- DOI: 10.1093/hmg/ddy118
Metabolomic studies identify changes in transmethylation and polyamine metabolism in a brain-specific mouse model of tuberous sclerosis complex
Abstract
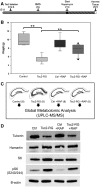
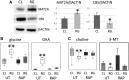
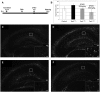
Tuberous sclerosis complex (TSC) is an autosomal dominant neurodevelopmental disorder and the quintessential disorder of mechanistic Target of Rapamycin Complex 1 (mTORC1) dysregulation. Loss of either causative gene, TSC1 or TSC2, leads to constitutive mTORC1 kinase activation and a pathologically anabolic state of macromolecular biosynthesis. Little is known about the organ-specific metabolic reprogramming that occurs in TSC-affected organs. Using a mouse model of TSC in which Tsc2 is disrupted in radial glial precursors and their neuronal and glial descendants, we performed an unbiased metabolomic analysis of hippocampi to identify Tsc2-dependent metabolic changes. Significant metabolic reprogramming was found in well-established pathways associated with mTORC1 activation, including redox homeostasis, glutamine/tricarboxylic acid cycle, pentose and nucleotide metabolism. Changes in two novel pathways were identified: transmethylation and polyamine metabolism. Changes in transmethylation included reduced methionine, cystathionine, S-adenosylmethionine (SAM-the major methyl donor), reduced SAM/S-adenosylhomocysteine ratio (cellular methylation potential), and elevated betaine, an alternative methyl donor. These changes were associated with alterations in SAM-dependent methylation pathways and expression of the enzymes methionine adenosyltransferase 2A and cystathionine beta synthase. We also found increased levels of the polyamine putrescine due to increased activity of ornithine decarboxylase, the rate-determining enzyme in polyamine synthesis. Treatment of Tsc2+/- mice with the ornithine decarboxylase inhibitor α-difluoromethylornithine, to reduce putrescine synthesis dose-dependently reduced hippocampal astrogliosis. These data establish roles for SAM-dependent methylation reactions and polyamine metabolism in TSC neuropathology. Importantly, both pathways are amenable to nutritional or pharmacologic therapy.
Figures




Similar articles
-
Ornithine decarboxylase, the rate-limiting enzyme of polyamine synthesis, modifies brain pathology in a mouse model of tuberous sclerosis complex.Hum Mol Genet. 2020 Aug 11;29(14):2395-2407. doi: 10.1093/hmg/ddaa121. Hum Mol Genet. 2020. PMID: 32588887 Free PMC article.
-
Upregulation of 6-phosphofructo-2-kinase (PFKFB3) by hyperactivated mammalian target of rapamycin complex 1 is critical for tumor growth in tuberous sclerosis complex.IUBMB Life. 2020 May;72(5):965-977. doi: 10.1002/iub.2232. Epub 2020 Jan 20. IUBMB Life. 2020. PMID: 31958214
-
Loss of Tsc2 in radial glia models the brain pathology of tuberous sclerosis complex in the mouse.Hum Mol Genet. 2009 Apr 1;18(7):1252-65. doi: 10.1093/hmg/ddp025. Epub 2009 Jan 15. Hum Mol Genet. 2009. PMID: 19150975 Free PMC article.
-
Pleiotropic effects of methionine adenosyltransferases deregulation as determinants of liver cancer progression and prognosis.J Hepatol. 2013 Oct;59(4):830-41. doi: 10.1016/j.jhep.2013.04.031. Epub 2013 May 7. J Hepatol. 2013. PMID: 23665184 Review.
-
Tuberous Sclerosis Complex.Keio J Med. 2025 Mar 25;74(1):42-51. doi: 10.2302/kjm.2023-0011-IR. Epub 2023 Aug 2. Keio J Med. 2025. PMID: 37532517 Review.
Cited by
-
Altered Metabolic Signaling and Potential Therapies in Polyglutamine Diseases.Metabolites. 2024 May 31;14(6):320. doi: 10.3390/metabo14060320. Metabolites. 2024. PMID: 38921455 Free PMC article. Review.
-
Metabolic Targets for Treatment of Autoimmune Diseases.Immunometabolism. 2020;2(2):e200012. doi: 10.20900/immunometab20200012. Epub 2020 Mar 31. Immunometabolism. 2020. PMID: 32341806 Free PMC article.
-
Regulation and metabolic functions of mTORC1 and mTORC2.Physiol Rev. 2021 Jul 1;101(3):1371-1426. doi: 10.1152/physrev.00026.2020. Epub 2021 Feb 18. Physiol Rev. 2021. PMID: 33599151 Free PMC article. Review.
-
Renal Transcriptome and Metabolome in Mice with Principal Cell-Specific Ablation of the Tsc1 Gene: Derangements in Pathways Associated with Cell Metabolism, Growth and Acid Secretion.Int J Mol Sci. 2022 Sep 13;23(18):10601. doi: 10.3390/ijms231810601. Int J Mol Sci. 2022. PMID: 36142537 Free PMC article.
-
Ornithine decarboxylase, the rate-limiting enzyme of polyamine synthesis, modifies brain pathology in a mouse model of tuberous sclerosis complex.Hum Mol Genet. 2020 Aug 11;29(14):2395-2407. doi: 10.1093/hmg/ddaa121. Hum Mol Genet. 2020. PMID: 32588887 Free PMC article.
References
-
- Crino P.B., Nathanson K.L., Henske E.P. (2006) The tuberous sclerosis complex. N. Engl. J. Med., 355, 1345–1356. - PubMed
-
- Consortium. (1993) Identification and characterization of the tuberous sclerosis gene on chromosome 16. Cell, 75, 1305–1315. - PubMed
-
- van Slegtenhorst M., de Hoogt R., Hermans C., Nellist M., Janssen B., Verhoef S., Lindhout D., van den Ouweland A., Halley D., Young J.. et al. (1997) Identification of the tuberous sclerosis gene TSC1 on chromosome 9q34. Science, 277, 805–808. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

