Genomic architecture of haddock (Melanogrammus aeglefinus) shows expansions of innate immune genes and short tandem repeats
- PMID: 29636006
- PMCID: PMC5894186
- DOI: 10.1186/s12864-018-4616-y
Genomic architecture of haddock (Melanogrammus aeglefinus) shows expansions of innate immune genes and short tandem repeats
Abstract
Background: Increased availability of genome assemblies for non-model organisms has resulted in invaluable biological and genomic insight into numerous vertebrates, including teleosts. Sequencing of the Atlantic cod (Gadus morhua) genome and the genomes of many of its relatives (Gadiformes) demonstrated a shared loss of the major histocompatibility complex (MHC) II genes 100 million years ago. An improved version of the Atlantic cod genome assembly shows an extreme density of tandem repeats compared to other vertebrate genome assemblies. Highly contiguous assemblies are therefore needed to further investigate the unusual immune system of the Gadiformes, and whether the high density of tandem repeats found in Atlantic cod is a shared trait in this group.
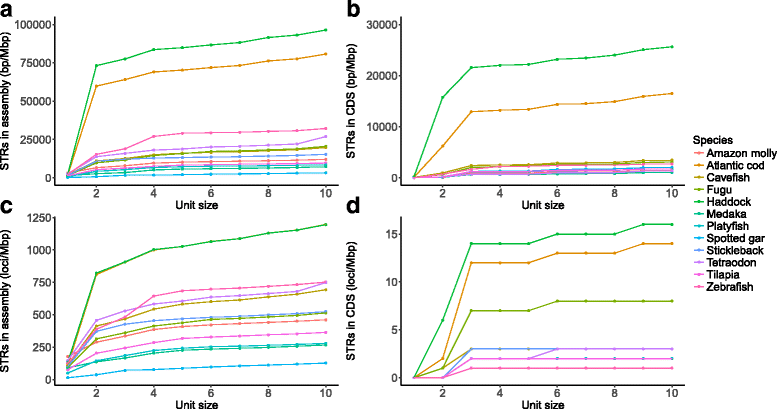
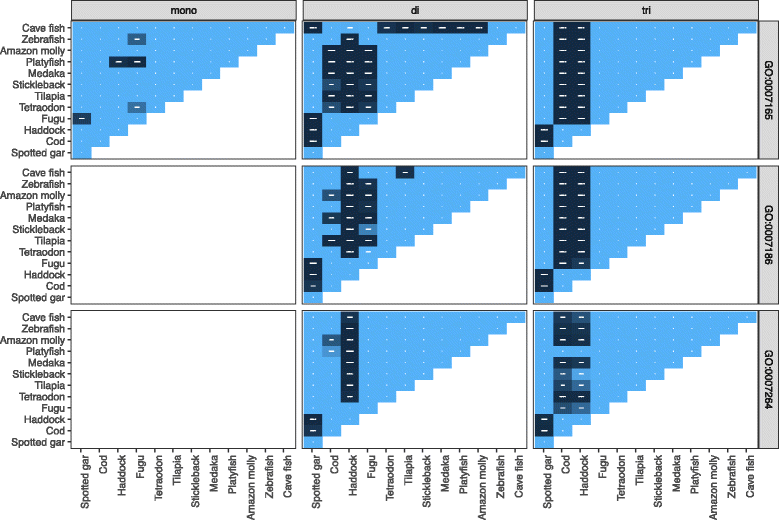
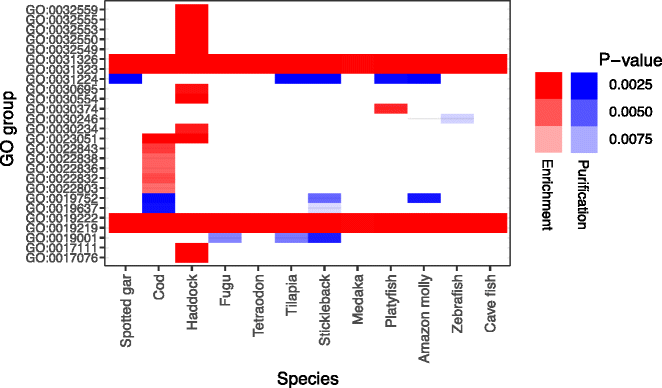
Results: Here, we have sequenced and assembled the genome of haddock (Melanogrammus aeglefinus) - a relative of Atlantic cod - using a combination of PacBio and Illumina reads. Comparative analyses reveal that the haddock genome contains an even higher density of tandem repeats outside and within protein coding sequences than Atlantic cod. Further, both species show an elevated number of tandem repeats in genes mainly involved in signal transduction compared to other teleosts. A characterization of the immune gene repertoire demonstrates a substantial expansion of MCHI in Atlantic cod compared to haddock. In contrast, the Toll-like receptors show a similar pattern of gene losses and expansions. For the NOD-like receptors (NLRs), another gene family associated with the innate immune system, we find a large expansion common to all teleosts, with possible lineage-specific expansions in zebrafish, stickleback and the codfishes.
Conclusions: The generation of a highly contiguous genome assembly of haddock revealed that the high density of short tandem repeats as well as expanded immune gene families is not unique to Atlantic cod - but possibly a feature common to all, or most, codfishes. A shared expansion of NLR genes in teleosts suggests that the NLRs have a more substantial role in the innate immunity of teleosts than other vertebrates. Moreover, we find that high copy number genes combined with variable genome assembly qualities may impede complete characterization of these genes, i.e. the number of NLRs in different teleost species might be underestimates.
Keywords: Atlantic cod; Genome assembly; Haddock; Microsatellites; NOD-like receptors; STRs.
Conflict of interest statement
Ethics approval
We have adhered to all local, national and international regulations and conventions, and we respected normal scientific ethical practices. The specimen used in this study comes from a wild population and was part of a larger haul of commercially fished individuals intended for human consumption. Following capture the fish was immediately stunned with a blunt object, then killed by bleeding, following standard procedure by local fishermen. Sampling in this manner does not fall under any specific legislation in Norway, but it is in accordance with the guidelines set by the ‘Norwegian consensus platform for replacement, reduction and refinement of animal experiments’ (
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

