Matrix metalloproteinase-9 (MMP9) is involved in the TNF-α-induced fusion of human M13SV1-Cre breast epithelial cells and human MDA-MB-435-pFDR1 cancer cells
- PMID: 29636110
- PMCID: PMC5894245
- DOI: 10.1186/s12964-018-0226-1
Matrix metalloproteinase-9 (MMP9) is involved in the TNF-α-induced fusion of human M13SV1-Cre breast epithelial cells and human MDA-MB-435-pFDR1 cancer cells
Abstract
Background: In addition to physiological events such as fertilisation, placentation, osteoclastogenesis, or tissue regeneration/wound healing, cell fusion is involved in pathophysiological conditions such as cancer. Cell fusion, which applies to both the proteins and conditions that induce the merging of two or more cells, is not a fully understood process. Inflammation/pro-inflammatory cytokines might be a positive trigger for cell fusion. Using a Cre-LoxP-based cell fusion assay we demonstrated that the fusion between human M13SV1-Cre breast epithelial cells and human MDA-MB-435-pFDR1 cancer cells was induced by the pro-inflammatory cytokine tumour necrosis factor-α (TNF-α).
Methods: The gene expression profile of the cells in the presence of TNF-α and under normoxic and hypoxic conditions was analysed by cDNA microarray analysis. cDNA microarray data were verified by qPCR, PCR, Western blot and zymography. Quantification of cell fusion events was determined by flow cytometry. Proteins of interest were either blocked or knocked-down using a specific inhibitor, siRNA or a blocking antibody.
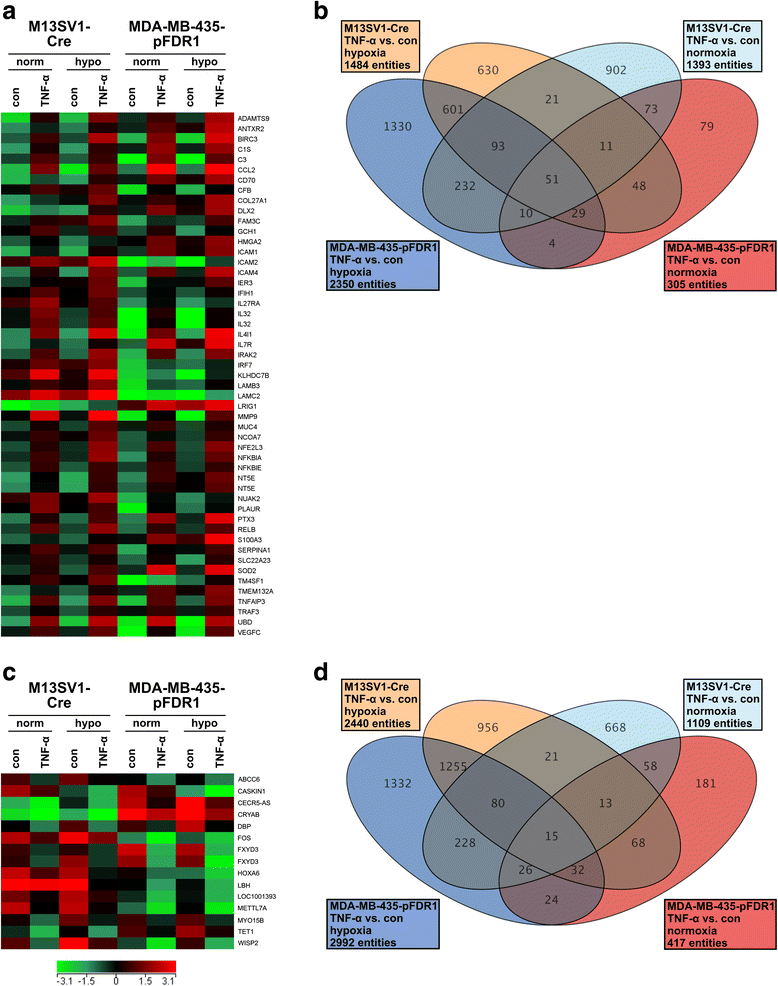
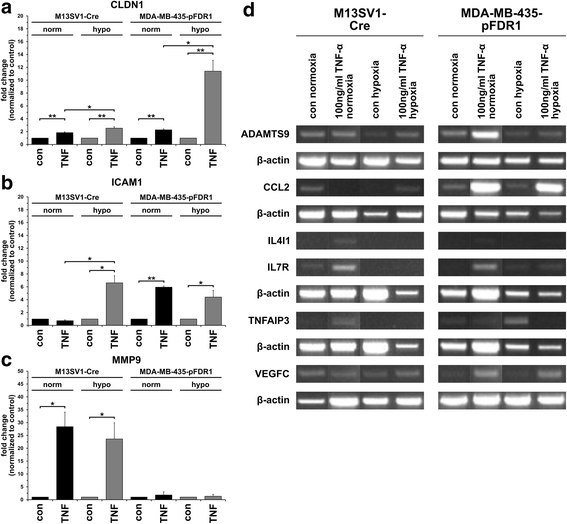
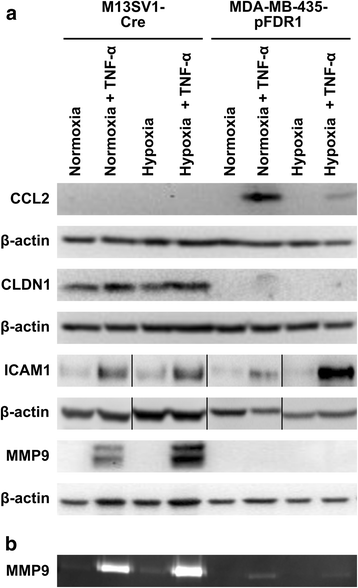
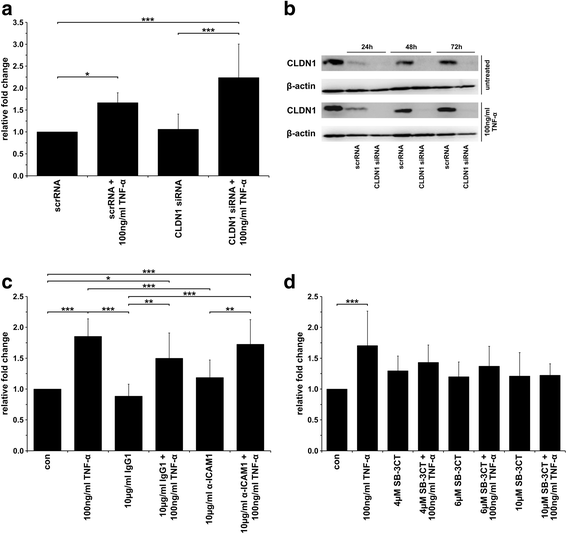
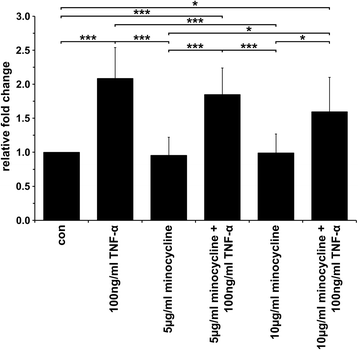
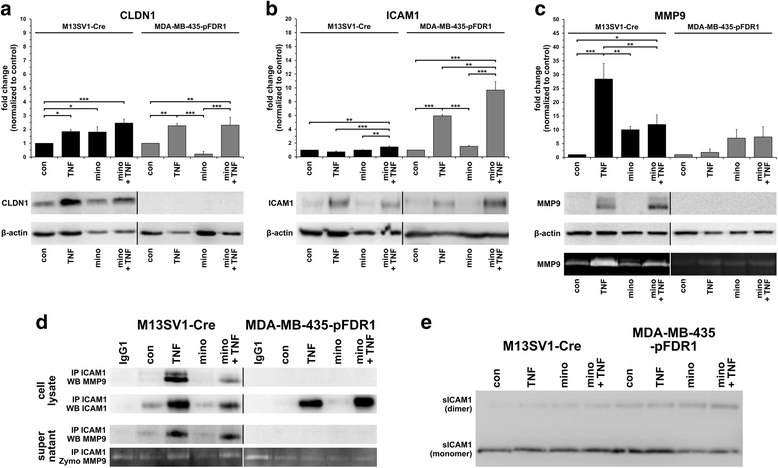
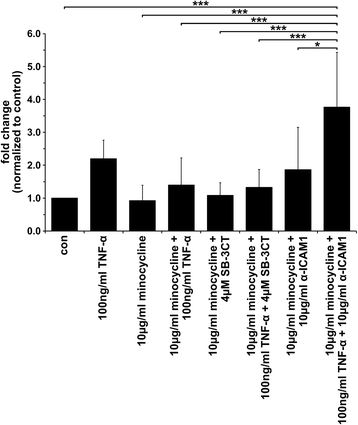
Results: The data showed an up-regulation of various genes, including claudin-1 (CLDN1), ICAM1, CCL2 and MMP9 in M13SV1-Cre and/or MDA-MB-435-pFDR1 cells. Inhibition of these proteins using a blocking ICAM1 antibody, CLDN1 siRNA or an MMP9 inhibitor showed that only the blockage of MMP9 was correlated with a decreased fusion rate of the cells. Likewise, the tetracycline-based antibiotic minocycline, which exhibits anti-inflammatory properties, was also effective in both inhibiting the TNF-α-induced MMP9 expression in M13SV1-Cre cells and blocking the TNF-α-induced fusion frequency of human M13SV1-Cre breast epithelial cells and human MDA-MB-435-pFDR1 cancer cells.
Conclusions: The matrix metalloproteinase-9 (MMP9) is most likely involved in the TNF-α-mediated fusion of human M13SV1-Cre breast epithelial cells and human MDA-MB-435-pFDR1 cancer cells. Likewise, our data indicate that the tetracycline-based antibiotic minocycline might exhibit anti-fusogenic properties because it inhibits a cell fusion-related mechanism.
Keywords: Breast cancer; Cell fusion; MMP9; Minocycline; TNF-α.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

