Massive mining of publicly available RNA-seq data from human and mouse
- PMID: 29636450
- PMCID: PMC5893633
- DOI: 10.1038/s41467-018-03751-6
Massive mining of publicly available RNA-seq data from human and mouse
Abstract
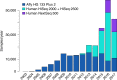
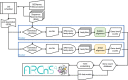
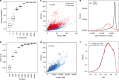
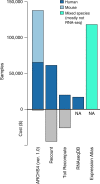
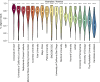
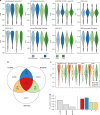
RNA sequencing (RNA-seq) is the leading technology for genome-wide transcript quantification. However, publicly available RNA-seq data is currently provided mostly in raw form, a significant barrier for global and integrative retrospective analyses. ARCHS4 is a web resource that makes the majority of published RNA-seq data from human and mouse available at the gene and transcript levels. For developing ARCHS4, available FASTQ files from RNA-seq experiments from the Gene Expression Omnibus (GEO) were aligned using a cloud-based infrastructure. In total 187,946 samples are accessible through ARCHS4 with 103,083 mouse and 84,863 human. Additionally, the ARCHS4 web interface provides intuitive exploration of the processed data through querying tools, interactive visualization, and gene pages that provide average expression across cell lines and tissues, top co-expressed genes for each gene, and predicted biological functions and protein-protein interactions for each gene based on prior knowledge combined with co-expression.
Conflict of interest statement
The authors declare no competing interests.
Figures
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

