A large-scale survey of the postmortem human microbiome, and its potential to provide insight into the living health condition
- PMID: 29636512
- PMCID: PMC5893548
- DOI: 10.1038/s41598-018-23989-w
A large-scale survey of the postmortem human microbiome, and its potential to provide insight into the living health condition
Abstract
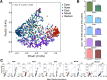
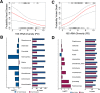
The microbiome plays many roles in human health, often through the exclusive lens of clinical interest. The inevitable end point for all living hosts, death, has its own altered microbiome configurations. However, little is understood about the ecology and changes of microbial communities after death, or their potential utility for understanding the health condition of the recently living. Here we reveal distinct postmortem microbiomes of human hosts from a large-scale survey of death cases representing a predominantly urban population, and demonstrated these microbiomes reflected antemortem health conditions within 24-48 hours of death. Our results characterized microbial community structure and predicted function from 188 cases representing a cross-section of an industrial-urban population. We found strong niche differentiation of anatomic habitat and microbial community turnover based on topographical distribution. Microbial community stability was documented up to two days after death. Additionally, we observed a positive relationship between cell motility and time since host death. Interestingly, we discovered evidence that microbial biodiversity is a predictor of antemortem host health condition (e.g., heart disease). These findings improve the understanding of postmortem host microbiota dynamics, and provide a robust dataset to test the postmortem microbiome as a tool for assessing health conditions in living populations.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Knight, R. et al. The Microbiome and Human Biology. Annu Rev Genomics Hum18, 10.1146/annurev-genom-083115-022438 (2017). - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

