Genome-Wide Association Study for Identification and Validation of Novel SNP Markers for Sr6 Stem Rust Resistance Gene in Bread Wheat
- PMID: 29636761
- PMCID: PMC5881291
- DOI: 10.3389/fpls.2018.00380
Genome-Wide Association Study for Identification and Validation of Novel SNP Markers for Sr6 Stem Rust Resistance Gene in Bread Wheat
Abstract
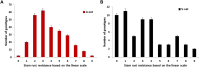
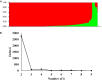
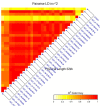
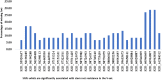
Stem rust (caused by Puccinia graminis f. sp. tritici Erikss. & E. Henn.), is a major disease in wheat (Triticum aestivium L.). However, in recent years it occurs rarely in Nebraska due to weather and the effective selection and gene pyramiding of resistance genes. To understand the genetic basis of stem rust resistance in Nebraska winter wheat, we applied genome-wide association study (GWAS) on a set of 270 winter wheat genotypes (A-set). Genotyping was carried out using genotyping-by-sequencing and ∼35,000 high-quality SNPs were identified. The tested genotypes were evaluated for their resistance to the common stem rust race in Nebraska (QFCSC) in two replications. Marker-trait association identified 32 SNP markers, which were significantly (Bonferroni corrected P < 0.05) associated with the resistance on chromosome 2D. The chromosomal location of the significant SNPs (chromosome 2D) matched the location of Sr6 gene which was expected in these genotypes based on pedigree information. A highly significant linkage disequilibrium (LD, r2 ) was found between the significant SNPs and the specific SSR marker for the Sr6 gene (Xcfd43). This suggests the significant SNP markers are tagging Sr6 gene. Out of the 32 significant SNPs, eight SNPs were in six genes that are annotated as being linked to disease resistance in the IWGSC RefSeq v1.0. The 32 significant SNP markers were located in nine haplotype blocks. All the 32 significant SNPs were validated in a set of 60 different genotypes (V-set) using single marker analysis. SNP markers identified in this study can be used in marker-assisted selection, genomic selection, and to develop KASP (Kompetitive Allele Specific PCR) marker for the Sr6 gene.
Highlights: Novel SNPs for Sr6 gene, an important stem rust resistant gene, were identified and validated in this study. These SNPs can be used to improve stem rust resistance in wheat.
Keywords: SNP validation; genome-wide association study; haplotypes; linkage disequilibrium; marker-assisted selection; single marker analysis.
Figures



References
-
- Baenziger P. S., Shelton D. R., Shipman M. J., Graybosch R. A. (2001). Breeding for end-use quality: reflections on the Nebraska experience. Euphytica 119 95–100. 10.1023/A:1017583514424 - DOI
-
- Belamkar V., Guttieri M. J., El-Basyoni I., Hussain W., Poland J., Jarquín D., et al. (2016). Integration of Genomic Selection in the Nebraska Wheat Breeding Program. Available at: https://pag.confex.com/pag/xxiv/webprogram/Paper21286.html [accessed January 25, 2018]. - PMC - PubMed
-
- Cingolani P., Platts A., Wang L. L., Coon M., Nguyen T., Wang L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, snpEFF: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6 80–92. 10.4161/fly.19695 - DOI - PMC - PubMed
Associated data
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

