Prenatally diagnosed distal 16p11.2 microdeletion with a novel association with congenital diaphragmatic hernia: a case report
- PMID: 29636920
- PMCID: PMC5889234
- DOI: 10.1002/ccr3.1369
Prenatally diagnosed distal 16p11.2 microdeletion with a novel association with congenital diaphragmatic hernia: a case report
Abstract
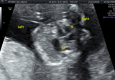
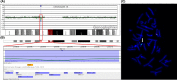
A prenatal case presenting with congenital diaphragmatic hernia (CDH) and distal 16p11.2 microdeletion suggests two possible causative hypotheses: (1) a functional effect of chromatin loopings between the distal and the proximal 16p11.2 microdeletion traits, associated with CHD; (2) a possible role of ATP2A1, a deleted gene involved in diaphragm development.
Keywords: 16p11.2 deletion syndrome; array‐CGH analysis; congenital diaphragmatic hernia; prenatal diagnosis; ultrasound fetal anomalies.
Figures
References
-
- Haroon, J. , and Chamberlain R. S.. 2013. An evidence‐based review of the current treatment of congenital diaphragmatic hernia. Clin. Pediatr. 52:115–124. - PubMed
-
- Zaiss, I. , Kehl S., Link K., Neff W., Schaible T., Sutterlin M., et al. 2011. Associated malformations in congenital diaphragmatic hernia. Am. J. Perinatol. 28:211–218. - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

