A Lexicon of DNA Modifications: Their Roles in Embryo Development and the Germline
- PMID: 29637072
- PMCID: PMC5880922
- DOI: 10.3389/fcell.2018.00024
A Lexicon of DNA Modifications: Their Roles in Embryo Development and the Germline
Abstract
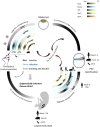
5-methylcytosine (5mC) on CpG dinucleotides has been viewed as the major epigenetic modification in eukaryotes for a long time. Apart from 5mC, additional DNA modifications have been discovered in eukaryotic genomes. Many of these modifications are thought to be solely associated with DNA damage. However, growing evidence indicates that some base modifications, namely 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), 5-carboxylcytosine (5caC), and N6-methadenine (6mA), may be of biological relevance, particularly during early stages of embryo development. Although abundance of these DNA modifications in eukaryotic genomes can be low, there are suggestions that they cooperate with other epigenetic markers to affect DNA-protein interactions, gene expression, defense of genome stability and epigenetic inheritance. Little is still known about their distribution in different tissues and their functions during key stages of the animal lifecycle. This review discusses current knowledge and future perspectives of these novel DNA modifications in the mammalian genome with a focus on their dynamic distribution during early embryonic development and their potential function in epigenetic inheritance through the germ line.
Keywords: 5hmC; 5mC; 6mA; embryo; epigenetic reprogramming; germ cells; modified bases in eukaryotic DNA.
Figures
References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

