Comparison of the oral microbiome in mouthwash and whole saliva samples
- PMID: 29641531
- PMCID: PMC5894969
- DOI: 10.1371/journal.pone.0194729
Comparison of the oral microbiome in mouthwash and whole saliva samples
Abstract
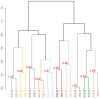
Population-based epidemiologic studies can provide important insight regarding the role of the microbiome in human health and disease. Buccal cells samples using commercial mouthwash have been obtained in large prospective cohorts for the purpose of studying human genomic DNA. We aimed to better understand if these mouthwash samples are also a valid resource for the study of the oral microbiome. We collected one saliva sample and one Scope mouthwash sample from 10 healthy subjects. Bacterial 16S rRNA genes from both types of samples were amplified, sequenced, and assigned to bacterial taxa. We comprehensively compared these paired samples for bacterial community composition and individual taxonomic abundance. We found that mouthwash samples yielded similar amount of bacterial DNA as saliva samples (p from Student's t-test for paired samples = 0.92). Additionally, the paired samples had similar within sample diversity (p from = 0.33 for richness, and p = 0.51 for Shannon index), and clustered as pairs for diversity when analyzed by unsupervised hierarchical cluster analysis. No significant difference was found in the paired samples with respect to the taxonomic abundance of major bacterial phyla, Bacteroidetes, Firmicutes, Proteobacteria, Fusobacteria, and Actinobacteria (FDR adjusted q values from Wilcoxin signed-rank test = 0.15, 0.15, 0.87, 1.00 and 0.15, respectively), and all identified genera, including genus Streptococcus (q = 0.21), Prevotella (q = 0.25), Neisseria (q = 0.37), Veillonella (q = 0.73), Fusobacterium (q = 0.19), and Porphyromonas (q = 0.60). These results show that mouthwash samples perform similarly to saliva samples for analysis of the oral microbiome. Mouthwash samples collected originally for analysis of human DNA are also a resource suitable for human microbiome research.
Conflict of interest statement
Figures


Similar articles
-
The Oral Microbiome in the Elderly With Dental Caries and Health.Front Cell Infect Microbiol. 2019 Jan 4;8:442. doi: 10.3389/fcimb.2018.00442. eCollection 2018. Front Cell Infect Microbiol. 2019. PMID: 30662876 Free PMC article.
-
Evaluation of Buccal Cell Samples for Studies of Oral Microbiota.Cancer Epidemiol Biomarkers Prev. 2017 Feb;26(2):249-253. doi: 10.1158/1055-9965.EPI-16-0538. Epub 2016 Oct 21. Cancer Epidemiol Biomarkers Prev. 2017. PMID: 27770008
-
Comparison of Oral Collection Methods for Studies of Microbiota.Cancer Epidemiol Biomarkers Prev. 2019 Jan;28(1):137-143. doi: 10.1158/1055-9965.EPI-18-0312. Epub 2018 Sep 27. Cancer Epidemiol Biomarkers Prev. 2019. PMID: 30262598 Free PMC article.
-
The oral microbiome and human health.J Oral Sci. 2017;59(2):201-206. doi: 10.2334/josnusd.16-0856. J Oral Sci. 2017. PMID: 28637979 Review.
-
Effects of chlorhexidine mouthwash on the oral microbiome.J Dent. 2021 Oct;113:103768. doi: 10.1016/j.jdent.2021.103768. Epub 2021 Aug 18. J Dent. 2021. PMID: 34418463 Review.
Cited by
-
Salivary Porphyromonas gingivalis predicts outcome in oral squamous cell carcinomas: a cohort study.BMC Oral Health. 2021 May 3;21(1):228. doi: 10.1186/s12903-021-01580-6. BMC Oral Health. 2021. PMID: 33941164 Free PMC article.
-
Oral Microbiome: A Review of Its Impact on Oral and Systemic Health.Microorganisms. 2024 Aug 29;12(9):1797. doi: 10.3390/microorganisms12091797. Microorganisms. 2024. PMID: 39338471 Free PMC article. Review.
-
Increasing Reproducibility in Oral Microbiome Research.Methods Mol Biol. 2021;2327:1-15. doi: 10.1007/978-1-0716-1518-8_1. Methods Mol Biol. 2021. PMID: 34410636
-
Systematic analyses uncover robust salivary microbial signatures and host-microbiome perturbations in oral squamous cell carcinoma.mSystems. 2025 Feb 18;10(2):e0124724. doi: 10.1128/msystems.01247-24. Epub 2025 Jan 28. mSystems. 2025. PMID: 39873508 Free PMC article.
-
Antiseptics: An expeditious third force in the prevention and management of coronavirus diseases.Curr Res Microb Sci. 2024 Oct 16;7:100293. doi: 10.1016/j.crmicr.2024.100293. eCollection 2024. Curr Res Microb Sci. 2024. PMID: 39497935 Free PMC article. Review.
References
-
- Yamashita Y, Takeshita T. The oral microbiome and human health. J Oral Sci. 2017;59(2):201–6. doi: 10.2334/josnusd.16-0856 . - DOI - PubMed
-
- Ohlrich EJ, Cullinan MP, Leichter JW. Diabetes, periodontitis, and the subgingival microbiota. J Oral Microbiol. 2010;2 doi: 10.3402/jom.v2i0.5818 . - DOI - PMC - PubMed
-
- Koren O, Spor A, Felin J, Fak F, Stombaugh J, Tremaroli V, et al. Human oral, gut, and plaque microbiota in patients with atherosclerosis. Proc Natl Acad Sci U S A. 2011;108 Suppl 1:4592–8. doi: 10.1073/pnas.1011383107 . - DOI - PMC - PubMed
-
- Ahn J, Chen CY, Hayes RB. Oral microbiome and oral and gastrointestinal cancer risk. Cancer causes & control: CCC. 2012;23(3):399–404. doi: 10.1007/s10552-011-9892-7 . - DOI - PMC - PubMed
-
- Fan X, Alekseyenko AV, Wu J, Peters BA, Jacobs EJ, Gapstur SM, et al. Human oral microbiome and prospective risk for pancreatic cancer: a population-based nested case-control study. Gut. 2018;67(1):120–7. doi: 10.1136/gutjnl-2016-312580 . - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

