Use of capillary Western immunoassay (Wes) for quantification of dystrophin levels in skeletal muscle of healthy controls and individuals with Becker and Duchenne muscular dystrophy
- PMID: 29641567
- PMCID: PMC5895072
- DOI: 10.1371/journal.pone.0195850
Use of capillary Western immunoassay (Wes) for quantification of dystrophin levels in skeletal muscle of healthy controls and individuals with Becker and Duchenne muscular dystrophy
Erratum in
-
Correction: Use of capillary Western immunoassay (Wes) for quantification of dystrophin levels in skeletal muscle of healthy controls and individuals with Becker and Duchenne muscular dystrophy.PLoS One. 2025 Nov 13;20(11):e0336651. doi: 10.1371/journal.pone.0336651. eCollection 2025. PLoS One. 2025. PMID: 41231766 Free PMC article.
Abstract
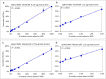
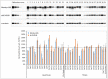
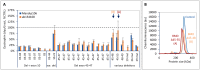
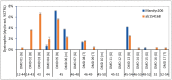
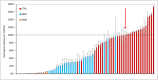
Duchenne muscular dystrophy (DMD) is a neuromuscular disease characterized by progressive weakness of the skeletal and cardiac muscles. This X-linked disorder is caused by open reading frame disrupting mutations in the DMD gene, resulting in strong reduction or complete absence of dystrophin protein. In order to use dystrophin as a supportive or even surrogate biomarker in clinical studies on investigational drugs aiming at correcting the primary cause of the disease, the ability to reliably quantify dystrophin expression in muscle biopsies of DMD patients pre- and post-treatment is essential. Here we demonstrate the application of the ProteinSimple capillary immunoassay (Wes) method, a gel- and blot-free method requiring less sample, antibody and time to run than conventional Western blot assay. We optimized dystrophin quantification by Wes using 2 different antibodies and found it to be highly sensitive, reproducible and quantitative over a large dynamic range. Using a healthy control muscle sample as a reference and α-actinin as a protein loading/muscle content control, a panel of skeletal muscle samples consisting of 31 healthy controls, 25 Becker Muscle dystrophy (BMD) and 17 DMD samples was subjected to Wes analysis. In healthy controls dystrophin levels varied 3 to 5-fold between the highest and lowest muscle samples, with the reference sample representing the average of all 31 samples. In BMD muscle samples dystrophin levels ranged from 10% to 90%, with an average of 33% of the healthy muscle average, while for the DMD samples the average dystrophin level was 1.3%, ranging from 0.7% to 7% of the healthy muscle average. In conclusion, Wes is a suitable, efficient and reliable method for quantification of dystrophin expression as a biomarker in DMD clinical drug development.
Conflict of interest statement
Figures



References
-
- Emery A. Duchenne muscular dystrophy. 2nd ed Oxford, England: Oxford University Press; 1993.
-
- Ryder S, Leadley R, Armstrong N, Westwood M, de Kock S, Butt T, et al. The burden, epidemiology, costs and treatment for Duchenne muscular dystrophy: an evidence review. Orphanet J Rare Dis. 2017;12: 79 doi: 10.1186/s13023-017-0631-3 - DOI - PMC - PubMed
-
- Muntoni F, Torelli S, Ferlini A. Dystrophin and mutations: one gene, several proteins, multiple phenotypes. Lancet Neurol. 2003;2: 731–740. - PubMed
-
- Abbs S, Tuffery-Giraud S, Bakker E, Ferlini A, Sejersen T, Mueller C. Best practice guidelines on molecular diagnostics in Duchenne/Becker muscular dystrophies. Neuromuscul Disord. 2010;20: 422–427. doi: 10.1016/j.nmd.2010.04.005 - DOI - PubMed
-
- Allen D, Whitehead N, Froehner S. Absence of dystrophin disrupts skeletal muscle signaling: Roles of Ca2+, reactive oxygen spcies, and nitric oxide in the development of muscular dystrophy. Physiol Rev. 2016;96: 253–305. doi: 10.1152/physrev.00007.2015 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

