FOXF1 Inhibits Pulmonary Fibrosis by Preventing CDH2-CDH11 Cadherin Switch in Myofibroblasts
- PMID: 29642003
- PMCID: PMC5947867
- DOI: 10.1016/j.celrep.2018.03.067
FOXF1 Inhibits Pulmonary Fibrosis by Preventing CDH2-CDH11 Cadherin Switch in Myofibroblasts
Abstract
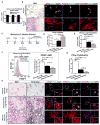
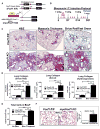
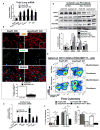
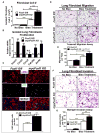
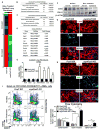
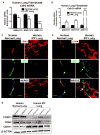
Idiopathic pulmonary fibrosis (IPF) is characterized by aberrant accumulation of collagen-secreting myofibroblasts. Development of effective therapies is limited due to incomplete understanding of molecular mechanisms regulating myofibroblast expansion. FOXF1 transcription factor is expressed in resident lung fibroblasts, but its role in lung fibrosis remains unknown due to the lack of genetic mouse models. Through comprehensive analysis of human IPF genomics data, lung biopsies, and transgenic mice with fibroblast-specific inactivation of FOXF1, we show that FOXF1 inhibits pulmonary fibrosis. FOXF1 deletion increases myofibroblast invasion and collagen secretion and promotes a switch from N-cadherin (CDH2) to Cadherin-11 (CDH11), which is a critical step in the acquisition of the pro-fibrotic phenotype. FOXF1 directly binds to Cdh2 and Cdh11 promoters and differentially regulates transcription of these genes. Re-expression of CDH2 or inhibition of CDH11 in FOXF1-deficient cells reduces myofibroblast invasion in vitro. FOXF1 inhibits pulmonary fibrosis by regulating a switch from CDH2 to CDH11 in lung myofibroblasts.
Keywords: CDH11; CDH2; FOXF1; cadherins; myofibroblasts; pulmonary fibrosis; transgenic mice.
Copyright © 2018 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Agarwal SK, Lee DM, Kiener HP, Brenner MB. Coexpression of two mesenchymal cadherins, cadherin 11 and N-cadherin, on murine fibroblast-like synoviocytes. Arthritis Rheum. 2008;58:1044–1054. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

