Crystal Structure of the Isocitrate Dehydrogenase 2 from Acinetobacter baumannii (AbIDH2) Reveals a Novel Dimeric Structure with Two Monomeric-IDH-Like Subunits
- PMID: 29642588
- PMCID: PMC5979607
- DOI: 10.3390/ijms19041131
Crystal Structure of the Isocitrate Dehydrogenase 2 from Acinetobacter baumannii (AbIDH2) Reveals a Novel Dimeric Structure with Two Monomeric-IDH-Like Subunits
Abstract
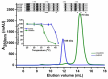
Monomeric isocitrate dehydrogenases (IDHs) have a single polypeptide sizing around 85 kDa. The IDH2 from the opportunistic bacterium Acinetobacter baumannii (AbIDH2) with a mass of 83 kDa was formerly recognized as a typical monomeric IDH. However, both size exclusion chromatography and analytical ultracentrifugation analysis indicated that AbIDH2 exists as a homodimer in solution. The crystallographic study of the substrate/coenzyme-free AbIDH2 gave a dimeric structure and each subunit contained a domain I and a domain II. The dimeric assembly is mainly stabilized by hydrophobic interactions (16 hydrogen bonds and 11 salt bridges) from the dimer's interface platform, which centered around the three parallel helices (α4, α12, and α17) and one loop from the domain II. Kinetic analysis showed that the dimeric AbIDH2 showed much lower catalytic efficiency (0.39 μM-1·s-1) as compared to the typical monomeric IDHs (~15 μM-1·s-1). Key residues crucial for dimer formation were simultaneously changed to generate the mutant mAbIDH2. The disruption of the hydrophobic forces disassociated the dimeric AbIDH2, making mAbIDH2 a monomeric enzyme. mAbIDH2 sustained specific activity (21.9 ± 2 U/mg) comparable to AbIDH2 (25.4 ± 0.7 U/mg). However, mAbIDH2 proved to be a thermolabile enzyme, indicating that the thermostable dimeric AbIDH2 may have a physiological significance for the growth and pathogenesis of A. baumannii. Phylogenetic analysis demonstrated the existence of numerous AbIDH2 homologous proteins, thus expanding the monomeric IDH protein family.
Keywords: Acinetobacter baumannii; crystal structure; dimerization; isocitrate dehydrogenase; phylogenetic studies; thermostability.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Jo S.H., Son M.K., Koh H.J., Lee S.M., Song I.H., Kim Y.O., Lee Y.S., Jeong K.S., Kim W.B., Park J.W., et al. Control of mitochondrial redox balance and cellular defense against oxidative damage by mitochondrial NADP+-dependent isocitrate dehydrogenase. J. Biol. Chem. 2001;276:16168–16176. doi: 10.1074/jbc.M010120200. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

