Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers
- PMID: 29642940
- PMCID: PMC5896039
- DOI: 10.1186/s40168-018-0451-2
Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers
Abstract
Background: Alterations of gut microbiota are associated with colorectal cancer (CRC) in different populations and several bacterial species were found to contribute to the tumorigenesis. The potential use of gut microbes as markers for early diagnosis has also been reported. However, cohort specific noises may distort the structure of microbial dysbiosis in CRC and lead to inconsistent results among studies. In this regard, our study targeted at exploring changes in gut microbiota that are universal across populations at species level.
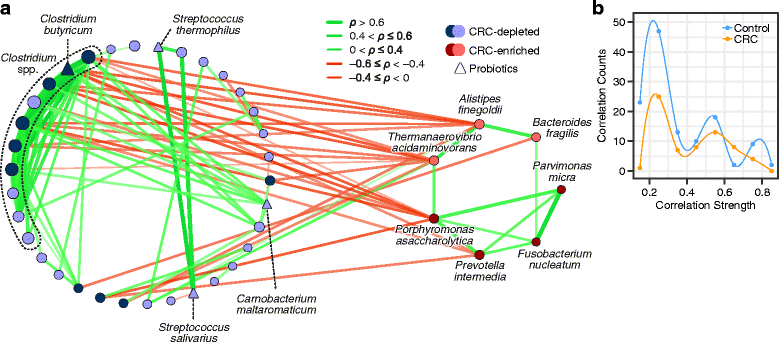
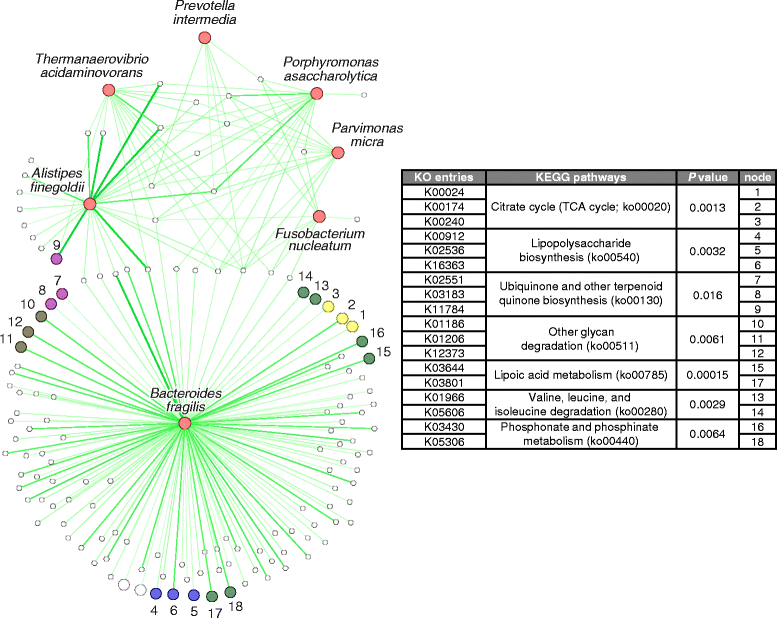
Results: Based on the combined analysis of 526 metagenomic samples from Chinese, Austrian, American, and German and French cohorts, seven CRC-enriched bacteria (Bacteroides fragilis, Fusobacterium nucleatum, Porphyromonas asaccharolytica, Parvimonas micra, Prevotella intermedia, Alistipes finegoldii, and Thermanaerovibrio acidaminovorans) have been identified across populations. The seven enriched bacterial markers classified cases from controls with an area under the receiver-operating characteristics curve (AUC) of 0.80 across the different populations. Abundance correlation analysis demonstrated that CRC-enriched and CRC-depleted bacteria respectively formed their own mutualistic networks, in which the latter was disjointed in CRC. The CRC-enriched bacteria have been found to be correlated with lipopolysaccharide and energy biosynthetic pathways.
Conclusions: Our study identified potential diagnostic bacterial markers that are robust across populations, indicating their potential universal use for non-invasive CRC diagnosis. We also elucidated the ecological networks and functional capacities of CRC-associated microbiota.
Keywords: Colorectal cancer; Diagnostic marker; Ecology; Microbiota.
Conflict of interest statement
Ethics approval and consent to participate
The study conformed to the ethical principles outlined by the Declaration of Helsinki and was approved by the Institutional Review Boards of the Chinese University of Hong Kong.
Consent for publication
Not applicable
Competing interests
The authors declare that they have no competing interest.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures


References
-
- Zhao LY, Zhang X, Zuo T, Yu J. The composition of colonic commensal Bacteria according to anatomical localization in colorectal Cancer. Engineering. 2017;3(1):90–97. doi: 10.1016/J.ENG.2017.01.012. - DOI
-
- Kostic AD, Chun EY, Robertson L, Glickman JN, Gallini CA, Michaud M, Clancy TE, Chung DC, Lochhead P, Hold GL, et al. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe. 2013;14(2):207–215. doi: 10.1016/j.chom.2013.07.007. - DOI - PMC - PubMed
-
- Goodwin AC, Shields CED, Wu SG, Huso DL, Wu XQ, Murray-Stewart TR, Hacker-Prietz A, Rabizadeh S, Woster PM, Sears CL, et al. Polyamine catabolism contributes to enterotoxigenic Bacteroides fragilis-induced colon tumorigenesis. P Natl Acad Sci USA. 2011;108(37):15354–15359. doi: 10.1073/pnas.1010203108. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

