Whole-Genome Sequencing of Recent Listeria monocytogenes Isolates from Germany Reveals Population Structure and Disease Clusters
- PMID: 29643197
- PMCID: PMC5971532
- DOI: 10.1128/JCM.00119-18
Whole-Genome Sequencing of Recent Listeria monocytogenes Isolates from Germany Reveals Population Structure and Disease Clusters
Abstract
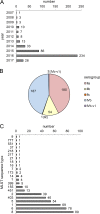
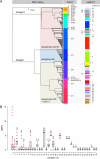
Listeria monocytogenes causes foodborne outbreaks with high mortality. For improvement of outbreak cluster detection, the German consiliary laboratory for listeriosis implemented whole-genome sequencing (WGS) in 2015. A total of 424 human L. monocytogenes isolates collected in 2007 to 2017 were subjected to WGS and core-genome multilocus sequence typing (cgMLST). cgMLST grouped the isolates into 38 complexes, reflecting 4 known and 34 unknown disease clusters. Most of these complexes were confirmed by single nucleotide polymorphism (SNP) calling, but some were further differentiated. Interestingly, several cgMLST cluster types were further subtyped by pulsed-field gel electrophoresis, partly due to phage insertions in the accessory genome. Our results highlight the usefulness of cgMLST for routine cluster detection but also show that cgMLST complexes require validation by methods providing higher typing resolution. Twelve cgMLST clusters included recent cases, suggesting activity of the source. Therefore, the cgMLST nomenclature data presented here may support future public health actions.
Keywords: core genome; listeriosis; outbreak detection; pathogen surveillance; subtyping.
Copyright © 2018 American Society for Microbiology.
Figures




Similar articles
-
Whole genome sequencing analyses of Listeria monocytogenes that persisted in a milkshake machine for a year and caused illnesses in Washington State.BMC Microbiol. 2017 Jun 15;17(1):134. doi: 10.1186/s12866-017-1043-1. BMC Microbiol. 2017. PMID: 28619007 Free PMC article.
-
Retrospective investigation of listeriosis outbreaks in small ruminants using different analytical approaches for whole genome sequencing-based typing of Listeria monocytogenes.Infect Genet Evol. 2020 Jan;77:104047. doi: 10.1016/j.meegid.2019.104047. Epub 2019 Oct 17. Infect Genet Evol. 2020. PMID: 31629888
-
Defining and Evaluating a Core Genome Multilocus Sequence Typing Scheme for Whole-Genome Sequence-Based Typing of Listeria monocytogenes.J Clin Microbiol. 2015 Sep;53(9):2869-76. doi: 10.1128/JCM.01193-15. Epub 2015 Jul 1. J Clin Microbiol. 2015. PMID: 26135865 Free PMC article.
-
Listeria monocytogenes: Strain Heterogeneity, Methods, and Challenges of Subtyping.J Food Sci. 2015 Dec;80(12):M2868-78. doi: 10.1111/1750-3841.13133. Epub 2015 Nov 20. J Food Sci. 2015. PMID: 26588067 Review.
-
Serotype to genotype: The changing landscape of listeriosis outbreak investigations.Food Microbiol. 2018 Oct;75:18-27. doi: 10.1016/j.fm.2017.06.013. Epub 2017 Jun 18. Food Microbiol. 2018. PMID: 30056958 Review.
Cited by
-
Novel type of pilus associated with a Shiga-toxigenic E. coli hybrid pathovar conveys aggregative adherence and bacterial virulence.Emerg Microbes Infect. 2018 Dec 5;7(1):203. doi: 10.1038/s41426-018-0209-8. Emerg Microbes Infect. 2018. PMID: 30514915 Free PMC article.
-
Isolate-Based Surveillance of Listeria monocytogenes by Whole Genome Sequencing in Austria.Front Microbiol. 2019 Oct 1;10:2282. doi: 10.3389/fmicb.2019.02282. eCollection 2019. Front Microbiol. 2019. PMID: 31632381 Free PMC article.
-
The Saprophytic Lifestyle of Listeria monocytogenes and Entry Into the Food-Processing Environment.Front Microbiol. 2022 Mar 8;13:789801. doi: 10.3389/fmicb.2022.789801. eCollection 2022. Front Microbiol. 2022. PMID: 35350628 Free PMC article. Review.
-
Genome Typing and Epidemiology of Human Listeriosis in New Zealand, 1999 to 2018.J Clin Microbiol. 2021 Oct 19;59(11):e0084921. doi: 10.1128/JCM.00849-21. Epub 2021 Aug 18. J Clin Microbiol. 2021. PMID: 34406797 Free PMC article.
-
Status and potential of bacterial genomics for public health practice: a scoping review.Implement Sci. 2019 Aug 13;14(1):79. doi: 10.1186/s13012-019-0930-2. Implement Sci. 2019. PMID: 31409417 Free PMC article.
References
-
- European Center for Disease Control. 2016. Annual epidemiological report 2016—listeriosis. European Center for Disease Control, Stockholm, Sweden: https://ecdc.europa.eu/sites/portal/files/documents/Listeriosis%20-%20An....
-
- Barton Behravesh C, Jones TF, Vugia DJ, Long C, Marcus R, Smith K, Thomas S, Zansky S, Fullerton KE, Henao OL, Scallan E, FoodNet Working G . 2011. Deaths associated with bacterial pathogens transmitted commonly through food: foodborne diseases active surveillance network (FoodNet), 1996–2005. J Infect Dis 204:263–267. doi:10.1093/infdis/jir263. - DOI - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

