Truncated CPSF6 Forms Higher-Order Complexes That Bind and Disrupt HIV-1 Capsid
- PMID: 29643241
- PMCID: PMC6002704
- DOI: 10.1128/JVI.00368-18
Truncated CPSF6 Forms Higher-Order Complexes That Bind and Disrupt HIV-1 Capsid
Abstract
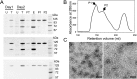
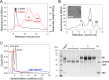
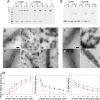
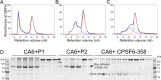
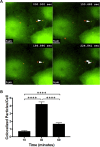
Cleavage and polyadenylation specificity factor 6 (CPSF6) is a human protein that binds HIV-1 capsid and mediates nuclear transport and integration targeting of HIV-1 preintegration complexes. Truncation of the protein at its C-terminal nuclear-targeting arginine/serine-rich (RS) domain produces a protein, CPSF6-358, that potently inhibits HIV-1 infection by targeting the capsid and inhibiting nuclear entry. To understand the molecular mechanism behind this restriction, the interaction between CPSF6-358 and HIV-1 capsid was characterized using in vitro and in vivo assays. Purified CPSF6-358 protein formed oligomers and bound in vitro-assembled wild-type (WT) capsid protein (CA) tubes, but not CA tubes containing a mutation in the putative binding site of CPSF6. Intriguingly, binding of CPSF6-358 oligomers to WT CA tubes physically disrupted the tubular assemblies into small fragments. Furthermore, fixed- and live-cell imaging showed that stably expressed CPSF6-358 forms cytoplasmic puncta upon WT HIV-1 infection and leads to capsid permeabilization. These events did not occur when the HIV-1 capsid contained a mutation known to prevent CPSF6 binding, nor did they occur in the presence of a small-molecule inhibitor of capsid binding to CPSF6-358. Together, our in vitro biochemical and transmission electron microscopy data and in vivo intracellular imaging results provide the first direct evidence for an oligomeric nature of CPSF6-358 and suggest a plausible mechanism for restriction of HIV-1 infection by CPSF6-358.IMPORTANCE After entry into cells, the HIV-1 capsid, which contains the viral genome, interacts with numerous host cell factors to facilitate crucial events required for replication, including uncoating. One such host cell factor, called CPSF6, is predominantly located in the cell nucleus and interacts with HIV-1 capsid. The interaction between CA and CPSF6 is critical during HIV-1 replication in vivo Truncation of CPSF6 leads to its localization to the cell cytoplasm and inhibition of HIV-1 infection. Here, we determined that truncated CPSF6 protein forms large higher-order complexes that bind directly to HIV-1 capsid, leading to its disruption. Truncated CPSF6 expression in cells leads to premature capsid uncoating that is detrimental to HIV-1 infection. Our study provides the first direct evidence for an oligomeric nature of truncated CPSF6 and insights into the highly regulated process of HIV-1 capsid uncoating.
Keywords: CPSF6; HIV; TEM; capsid; imaging; restriction.
Copyright © 2018 American Society for Microbiology.
Figures



Similar articles
-
HIV-1-induced translocation of CPSF6 to biomolecular condensates.Trends Microbiol. 2024 Aug;32(8):781-790. doi: 10.1016/j.tim.2024.01.001. Epub 2024 Jan 23. Trends Microbiol. 2024. PMID: 38267295 Free PMC article. Review.
-
CPSF6 promotes HIV-1 preintegration complex function.J Virol. 2025 May 20;99(5):e0049025. doi: 10.1128/jvi.00490-25. Epub 2025 Apr 9. J Virol. 2025. PMID: 40202316 Free PMC article.
-
Cytoplasmic CPSF6 Regulates HIV-1 Capsid Trafficking and Infection in a Cyclophilin A-Dependent Manner.mBio. 2021 Mar 23;12(2):e03142-20. doi: 10.1128/mBio.03142-20. mBio. 2021. PMID: 33758083 Free PMC article.
-
Analysis of CA Content and CPSF6 Dependence of Early HIV-1 Replication Complexes in SupT1-R5 Cells.mBio. 2019 Nov 5;10(6):e02501-19. doi: 10.1128/mBio.02501-19. mBio. 2019. PMID: 31690677 Free PMC article.
-
Role of Transportin-SR2 in HIV-1 Nuclear Import.Viruses. 2021 May 4;13(5):829. doi: 10.3390/v13050829. Viruses. 2021. PMID: 34064404 Free PMC article. Review.
Cited by
-
CA Mutation N57A Has Distinct Strain-Specific HIV-1 Capsid Uncoating and Infectivity Phenotypes.J Virol. 2019 Apr 17;93(9):e00214-19. doi: 10.1128/JVI.00214-19. Print 2019 May 1. J Virol. 2019. PMID: 30814280 Free PMC article.
-
Pharmacologic hyperstabilisation of the HIV-1 capsid lattice induces capsid failure.Elife. 2024 Feb 13;13:e83605. doi: 10.7554/eLife.83605. Elife. 2024. PMID: 38347802 Free PMC article.
-
HIV capsids: orchestrators of innate immune evasion, pathogenesis and pandemicity.J Gen Virol. 2025 Jan;106(1):002057. doi: 10.1099/jgv.0.002057. J Gen Virol. 2025. PMID: 39804283 Free PMC article. Review.
-
HIV-1-induced translocation of CPSF6 to biomolecular condensates.Trends Microbiol. 2024 Aug;32(8):781-790. doi: 10.1016/j.tim.2024.01.001. Epub 2024 Jan 23. Trends Microbiol. 2024. PMID: 38267295 Free PMC article. Review.
-
Molecular Determinants of PQBP1 Binding to the HIV-1 Capsid Lattice.J Mol Biol. 2024 Feb 15;436(4):168409. doi: 10.1016/j.jmb.2023.168409. Epub 2023 Dec 20. J Mol Biol. 2024. PMID: 38128824 Free PMC article.
References
-
- Pertel T, Hausmann S, Morger D, Zuger S, Guerra J, Lascano J, Reinhard C, Santoni FA, Uchil PD, Chatel L, Bisiaux A, Albert ML, Strambio-De-Castillia C, Mothes W, Pizzato M, Grutter MG, Luban J. 2011. TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472:361–365. doi:10.1038/nature09976. - DOI - PMC - PubMed
-
- Liu PT, Schenk M, Walker VP, Dempsey PW, Kanchanapoomi M, Wheelwright M, Vazirnia A, Zhang X, Steinmeyer A, Zugel U, Hollis BW, Cheng G, Modlin RL. 2009. Convergence of IL-1beta and VDR activation pathways in human TLR2/1-induced antimicrobial responses. PLoS One 4:e5810. doi:10.1371/journal.pone.0005810. - DOI - PMC - PubMed
-
- Liu C, Perilla JR, Ning J, Lu M, Hou G, Ramalho R, Himes BA, Zhao G, Bedwell GJ, Byeon I-J, Ahn J, Gronenborn AM, Prevelige PE, Rousso I, Aiken C, Polenova T, Schulten K, Zhang P. 2016. Cyclophilin A stabilizes the HIV-1 capsid through a novel non-canonical binding site. Nat Commun 7:10714. doi:10.1038/ncomms10714. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

