Canine NAPEPLD-associated models of human myelin disorders
- PMID: 29643404
- PMCID: PMC5895582
- DOI: 10.1038/s41598-018-23938-7
Canine NAPEPLD-associated models of human myelin disorders
Abstract
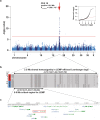
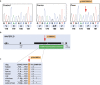
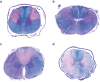
Canine leukoencephalomyelopathy (LEMP) is a juvenile-onset neurodegenerative disorder of the CNS white matter currently described in Rottweiler and Leonberger dogs. Genome-wide association study (GWAS) allowed us to map LEMP in a Leonberger cohort to dog chromosome 18. Subsequent whole genome re-sequencing of a Leonberger case enabled the identification of a single private homozygous non-synonymous missense variant located in the highly conserved metallo-beta-lactamase domain of the N-acyl phosphatidylethanolamine phospholipase D (NAPEPLD) gene, encoding an enzyme of the endocannabinoid system. We then sequenced this gene in LEMP-affected Rottweilers and identified a different frameshift variant, which is predicted to replace the C-terminal metallo-beta-lactamase domain of the wild type protein. Haplotype analysis of SNP array genotypes revealed that the frameshift variant was present in diverse haplotypes in Rottweilers, and also in Great Danes, indicating an old origin of this second NAPEPLD variant. The identification of different NAPEPLD variants in dog breeds affected by leukoencephalopathies with heterogeneous pathological features, implicates the NAPEPLD enzyme as important in myelin homeostasis, and suggests a novel candidate gene for myelination disorders in people.
Conflict of interest statement
Both the University of Minnesota and the University of Bern offer a genotyping test for LEMP, as well as several other genetic conditions in dogs, in their respective laboratories. None of the authors gain financially from the proceeds of these tests, which all go to support ongoing canine genetic research. Non-financial competing interests of authors from both Universities include informal advisory relationships concerning use and interpretation of genetic testing, as well as financial support for the research, from non-profit organizations that include the Leonberger Health Foundation (USA), the International Leonberger Union, and the Swiss and German Leonberger Clubs.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

