Intrinsically-disordered N-termini in human parechovirus 1 capsid proteins bind encapsidated RNA
- PMID: 29643409
- PMCID: PMC5895611
- DOI: 10.1038/s41598-018-23552-7
Intrinsically-disordered N-termini in human parechovirus 1 capsid proteins bind encapsidated RNA
Abstract
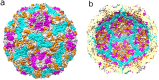
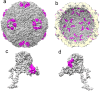
Human parechoviruses (HPeV) are picornaviruses with a highly-ordered RNA genome contained within icosahedrally-symmetric capsids. Ordered RNA structures have recently been shown to interact with capsid proteins VP1 and VP3 and facilitate virus assembly in HPeV1. Using an assay that combines reversible cross-linking, RNA affinity purification and peptide mass fingerprinting (RCAP), we mapped the RNA-interacting regions of the capsid proteins from the whole HPeV1 virion in solution. The intrinsically-disordered N-termini of capsid proteins VP1 and VP3, and unexpectedly, VP0, were identified to interact with RNA. Comparing these results to those obtained using recombinantly-expressed VP0 and VP1 confirmed the virion binding regions, and revealed unique RNA binding regions in the isolated VP0 not previously observed in the crystal structure of HPeV1. We used RNA fluorescence anisotropy to confirm the RNA-binding competency of each of the capsid proteins' N-termini. These findings suggests that dynamic interactions between the viral RNA and the capsid proteins modulate virus assembly, and suggest a novel role for VP0.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
The Structure of Human Parechovirus 1 Reveals an Association of the RNA Genome with the Capsid.J Virol. 2015 Nov 18;90(3):1377-86. doi: 10.1128/JVI.02346-15. Print 2016 Feb 1. J Virol. 2015. PMID: 26581987 Free PMC article.
-
Multiple capsid-stabilizing interactions revealed in a high-resolution structure of an emerging picornavirus causing neonatal sepsis.Nat Commun. 2016 Jul 20;7:11387. doi: 10.1038/ncomms11387. Nat Commun. 2016. PMID: 27435188 Free PMC article.
-
Structural Basis of Human Parechovirus Neutralization by Human Monoclonal Antibodies.J Virol. 2015 Sep;89(18):9571-80. doi: 10.1128/JVI.01429-15. Epub 2015 Jul 8. J Virol. 2015. PMID: 26157123 Free PMC article.
-
3D Puzzle at the Nanoscale-How do RNA Viruses Self-Assemble their Capsids into Perfectly Ordered Structures.Macromol Biosci. 2024 Sep;24(9):e2400088. doi: 10.1002/mabi.202400088. Epub 2024 Jun 24. Macromol Biosci. 2024. PMID: 38864315 Review.
-
How and why RNA genomes are (partially) ordered in viral capsids.Curr Opin Virol. 2022 Feb;52:203-210. doi: 10.1016/j.coviro.2021.11.014. Epub 2021 Dec 24. Curr Opin Virol. 2022. PMID: 34959081 Review.
Cited by
-
Perspectives on Viral RNA Genomes and the RNA Folding Problem.Viruses. 2020 Oct 5;12(10):1126. doi: 10.3390/v12101126. Viruses. 2020. PMID: 33027988 Free PMC article. Review.
-
Parechovirus A Pathogenesis and the Enigma of Genotype A-3.Viruses. 2019 Nov 14;11(11):1062. doi: 10.3390/v11111062. Viruses. 2019. PMID: 31739613 Free PMC article. Review.
-
Structural flexibility in the ordered domain of the dengue virus strain 2 capsid protein is critical for chaperoning viral RNA replication.Cell Mol Life Sci. 2025 Apr 28;82(1):184. doi: 10.1007/s00018-025-05712-x. Cell Mol Life Sci. 2025. PMID: 40293525 Free PMC article.
-
Capsid Structure of a Marine Algal Virus of the Order Picornavirales.J Virol. 2020 Apr 16;94(9):e01855-19. doi: 10.1128/JVI.01855-19. Print 2020 Apr 16. J Virol. 2020. PMID: 32024776 Free PMC article.
-
Prevalence and genetic diversity of Parechovirus.Virus Res. 2024 Nov;349:199461. doi: 10.1016/j.virusres.2024.199461. Epub 2024 Sep 18. Virus Res. 2024. PMID: 39278352 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

