Impact of a vegan diet on the human salivary microbiota
- PMID: 29643500
- PMCID: PMC5895596
- DOI: 10.1038/s41598-018-24207-3
Impact of a vegan diet on the human salivary microbiota
Abstract
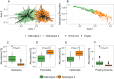
Little is known about the effect of long-term diet patterns on the composition and functional potential of the human salivary microbiota. In the present study, we sought to contribute to the ongoing elucidation of dietary effects on the oral microbial community by examining the diversity, composition and functional potential of the salivary microbiota in 160 healthy vegans and omnivores using 16S rRNA gene amplicon sequencing. We further sought to identify bacterial taxa in saliva associated with host inflammatory markers. We show that compositional differences in the salivary microbiota of vegans and omnivores is present at all taxonomic levels below phylum level and includes upper respiratory tract commensals (e.g. Neisseria subflava, Haemophilus parainfluenzae, and Rothia mucilaginosa) and species associated with periodontal disease (e.g. Campylobacter rectus and Porphyromonas endodontalis). Dietary intake of medium chain fatty acids, piscine mono- and polyunsaturated fatty acids, and dietary fibre was associated with bacterial diversity, community structure, as well as relative abundance of several species-level operational taxonomic units. Analysis of imputed genomic potential revealed several metabolic pathways differentially abundant in vegans and omnivores indicating possible effects of macro- and micro-nutrient intake. We also show that certain oral bacteria are associated with the systemic inflammatory state of the host.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Claesson, M. J. et al. Gut microbiota composition correlates with diet and health in the elderly. Nature488 (2012). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

