Genome evolution across 1,011 Saccharomyces cerevisiae isolates
- PMID: 29643504
- PMCID: PMC6784862
- DOI: 10.1038/s41586-018-0030-5
Genome evolution across 1,011 Saccharomyces cerevisiae isolates
Abstract
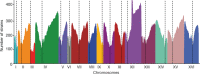
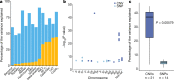
Large-scale population genomic surveys are essential to explore the phenotypic diversity of natural populations. Here we report the whole-genome sequencing and phenotyping of 1,011 Saccharomyces cerevisiae isolates, which together provide an accurate evolutionary picture of the genomic variants that shape the species-wide phenotypic landscape of this yeast. Genomic analyses support a single 'out-of-China' origin for this species, followed by several independent domestication events. Although domesticated isolates exhibit high variation in ploidy, aneuploidy and genome content, genome evolution in wild isolates is mainly driven by the accumulation of single nucleotide polymorphisms. A common feature is the extensive loss of heterozygosity, which represents an essential source of inter-individual variation in this mainly asexual species. Most of the single nucleotide polymorphisms, including experimentally identified functional polymorphisms, are present at very low frequencies. The largest numbers of variants identified by genome-wide association are copy-number changes, which have a greater phenotypic effect than do single nucleotide polymorphisms. This resource will guide future population genomics and genotype-phenotype studies in this classic model system.
Conflict of interest statement
The authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

