Mutation in Rice Abscisic Acid2 Results in Cell Death, Enhanced Disease-Resistance, Altered Seed Dormancy and Development
- PMID: 29643863
- PMCID: PMC5882781
- DOI: 10.3389/fpls.2018.00405
Mutation in Rice Abscisic Acid2 Results in Cell Death, Enhanced Disease-Resistance, Altered Seed Dormancy and Development
Abstract
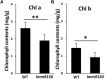
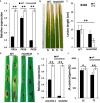
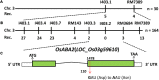
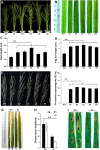
Lesion mimic mutants display spontaneous cell death, and thus are valuable for understanding the molecular mechanism of cell death and disease resistance. Although a lot of such mutants have been characterized in rice, the relationship between lesion formation and abscisic acid (ABA) synthesis pathway is not reported. In the present study, we identified a rice mutant, lesion mimic mutant 9150 (lmm9150), exhibiting spontaneous cell death, pre-harvest sprouting, enhanced growth, and resistance to rice bacterial and blast diseases. Cell death in the mutant was accompanied with excessive accumulation of H2O2. Enhanced disease resistance was associated with cell death and upregulation of defense-related genes. Map-based cloning identified a G-to-A point mutation resulting in a D-to-N substitution at the amino acid position 110 of OsABA2 (LOC_Os03g59610) in lmm9150. Knock-out of OsABA2 through CRISPR/Cas9 led to phenotypes similar to those of lmm9150. Consistent with the function of OsABA2 in ABA biosynthesis, ABA level in the lmm9150 mutant was significantly reduced. Moreover, exogenous application of ABA could rescue all the mutant phenotypes of lmm9150. Taken together, our data linked ABA deficiency to cell death and provided insight into the role of ABA in rice disease resistance.
Keywords: ABA/GA ratio; Oryza sativa L.; abscisic acid; defense responses; gibberellin; lesion mimic mutant; pre-harvest sprouting; xanthoxin dehydrogenase.
Figures



Similar articles
-
Functional characterization of xanthoxin dehydrogenase in rice.J Plant Physiol. 2014 Sep 1;171(14):1231-40. doi: 10.1016/j.jplph.2014.05.003. Epub 2014 Jun 2. J Plant Physiol. 2014. PMID: 25014258
-
Mutation of SPOTTED LEAF3 (SPL3) impairs abscisic acid-responsive signalling and delays leaf senescence in rice.J Exp Bot. 2015 Dec;66(22):7045-59. doi: 10.1093/jxb/erv401. Epub 2015 Aug 14. J Exp Bot. 2015. PMID: 26276867 Free PMC article.
-
OsABA3 is Crucial for Plant Survival and Resistance to Multiple Stresses in Rice.Rice (N Y). 2024 Jul 31;17(1):46. doi: 10.1186/s12284-024-00724-w. Rice (N Y). 2024. PMID: 39083143 Free PMC article.
-
Advances in the Genetic Basis and Molecular Mechanism of Lesion Mimic Formation in Rice.Plants (Basel). 2022 Aug 21;11(16):2169. doi: 10.3390/plants11162169. Plants (Basel). 2022. PMID: 36015472 Free PMC article. Review.
-
Rice Lesion Mimic Mutants (LMM): The Current Understanding of Genetic Mutations in the Failure of ROS Scavenging during Lesion Formation.Plants (Basel). 2021 Aug 4;10(8):1598. doi: 10.3390/plants10081598. Plants (Basel). 2021. PMID: 34451643 Free PMC article. Review.
Cited by
-
PROTEIN PHOSPHATASE 2C08, a Negative Regulator of Abscisic Acid Signaling, Promotes Internode Elongation in Rice.Int J Mol Sci. 2023 Jun 28;24(13):10821. doi: 10.3390/ijms241310821. Int J Mol Sci. 2023. PMID: 37445999 Free PMC article.
-
SnRK1α1 Antagonizes Cell Death Induced by Transient Overexpression of Arabidopsis thaliana ABI5 Binding Protein 2 (AFP2).Front Plant Sci. 2020 Sep 29;11:582208. doi: 10.3389/fpls.2020.582208. eCollection 2020. Front Plant Sci. 2020. PMID: 33133119 Free PMC article.
-
Brassinosteroids regulate rice seed germination through the BZR1-RAmy3D transcriptional module.Plant Physiol. 2022 May 3;189(1):402-418. doi: 10.1093/plphys/kiac043. Plant Physiol. 2022. PMID: 35139229 Free PMC article.
-
Seed Dormancy and Pre-Harvest Sprouting in Rice-An Updated Overview.Int J Mol Sci. 2021 Oct 30;22(21):11804. doi: 10.3390/ijms222111804. Int J Mol Sci. 2021. PMID: 34769234 Free PMC article. Review.
-
GWAS analysis of sorghum association panel lines identifies SNPs associated with disease response to Texas isolates of Colletotrichum sublineola.Theor Appl Genet. 2019 May;132(5):1389-1396. doi: 10.1007/s00122-019-03285-5. Epub 2019 Jan 28. Theor Appl Genet. 2019. PMID: 30688991
References
-
- Agrawal G. K., Yamazaki M., Kobayashi M., Hirochika R., Miyao A., Hirochika H. (2001). Screening of the rice viviparous mutants generated by endogenous retrotransposon Tos17 insertion. Tagging of a zeaxanthin epoxidase gene and a novel OsTATC gene. Plant Physiol. 125 1248–1257. 10.1104/pp.125.3.1248 - DOI - PMC - PubMed
-
- Anderson J., Badruzsaufari E., Schenk P. M., Manners J. M., Desmond O. J., Ehlert C. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16 3460–3479. 10.1105/tpc.104.025833 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

