Epigenetic versus Genetic Deregulation of the KEAP1/NRF2 Axis in Solid Tumors: Focus on Methylation and Noncoding RNAs
- PMID: 29643973
- PMCID: PMC5872633
- DOI: 10.1155/2018/2492063
Epigenetic versus Genetic Deregulation of the KEAP1/NRF2 Axis in Solid Tumors: Focus on Methylation and Noncoding RNAs
Abstract
Oxidative and electrophilic changes in cells are mainly coordinated by the KEAP1/NRF2 (Kelch-like erythroid-derived cap-n-collar homology- (ECH-) associated protein-1/nuclear factor (erythroid-derived 2)-like 2) axis. The physical interaction between these two proteins promotes the expression of several antioxidant defense genes in response to exogenous and endogenous insults. Recent studies demonstrated that KEAP1/NRF2 axis dysfunction is also strongly related to tumor progression and chemo- and radiotherapy resistance of cancer cells. In solid tumors, the KEAP1/NRF2 system is constitutively activated by the loss of KEAP1 or gain of NFE2L2 functions that leads to its nuclear accumulation and enhances the transcription of many cytoprotective genes. In addition to point mutations, epigenetic abnormalities, as aberrant promoter methylation, and microRNA (miRNA) and long noncoding RNA (lncRNA) deregulation were reported as emerging mechanisms of KEAP1/NRF2 axis modulation. This review will summarize the current knowledge about the epigenetic mechanisms that deregulate the KEAP1/NRF2 cascade in solid tumors and their potential usefulness as prognostic and predictive molecular markers.
Figures

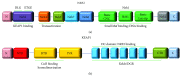
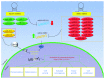
References
-
- Mcmahon M., Itoh K., Yamamoto M., Hayes J. D. Keap1-dependent proteasomal degradation of transcription factor Nrf2 contributes to the negative regulation of antioxidant response element-driven gene expression. The Journal of Biological Chemistry. 2003;278(24):21592–21600. doi: 10.1074/jbc.M300931200. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

