Molecular, phylogenetic and developmental analyses of Sall proteins in bilaterians
- PMID: 29644029
- PMCID: PMC5892016
- DOI: 10.1186/s13227-018-0096-z
Molecular, phylogenetic and developmental analyses of Sall proteins in bilaterians
Abstract
Background: Sall (Spalt-like) proteins are zinc-finger transcription factors involved in a number of biological processes. They have only been studied in a few model organisms, such as Drosophila melanogaster, Caenorhabditis elegans, Schmidtea mediterranea and some vertebrates. Further taxon sampling is critical to understand the evolution and diversification of this protein and its functional roles in animals.
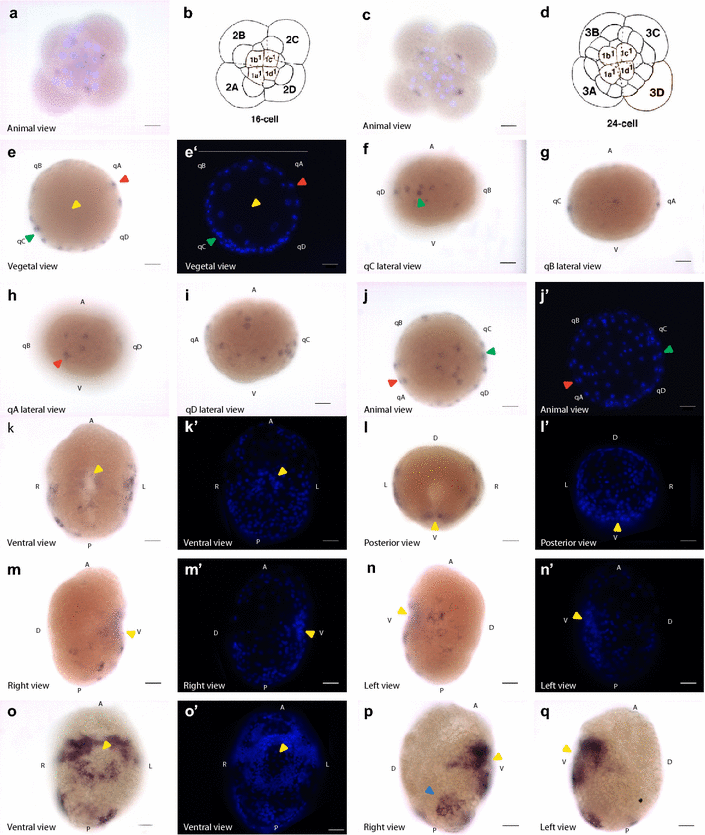
Results: Using genome and transcriptome mining, we confirmed the presence of sall genes in a range of additional animal taxa, for which their presence had not yet been described. We show that sall genes are broadly conserved across the Bilateria, and likely appeared in the bilaterian stem lineage. Our analysis of the protein domains shows that the characteristic arrangement of the multiple zinc-finger domains is conserved in bilaterians and may represent the ancient arrangement of this family of transcription factors. We also show the existence of a previously unknown zinc-finger domain. In situ hybridization was used to describe the gene expression patterns in embryonic and larval stages in two species of snails: Crepidula fornicata and Lottia gigantea. In L. gigantea, sall presents maternal expression, although later on the expression is restricted to the A and B quadrants during gastrulation and larval stage. In C. fornicata, sall has no maternal expression and it is expressed mainly in the A, C and D quadrants during blastula stages and in an asymmetric fashion during the larval stage.
Discussion: Our results suggest that the bilaterian common ancestor had a Sall protein with at least six zinc-finger domains. The evolution of Sall proteins in bilaterians might have occurred mostly as a result of the loss of protein domains and gene duplications leading to diversification. The new evidence complements previous studies in highlighting an important role of Sall proteins in bilaterian development. Our results show maternal expression of sall in the snail L. gigantea, but not C. fornicata. The asymmetric expression shown in the ectoderm of the trochophore larva of snails is probably related to shell/mantle development. The observed sall expression in cephalic tissue in snails and some other bilaterians suggests a possible ancestral role of sall in neural development in bilaterians.
Keywords: Crepidula fornicata; Gastropoda; Gene evolution; Lottia gigantea; Protein domains; Sall; Spalt; Spiralia.
Figures




References
-
- Kuhnlein RP, Frommer G, Friedrich M, Gonzalez-Gaitan M, Weber A, Wagner-Bernholz JF, Gehring WJ, Jackle H, Schuh R. Spalt encodes an evolutionarily conserved zinc finger protein of novel structure which provides homeotic gene function in the head and tail region of the Drosophila embryo. EMBO J. 1994;13(1):168–179. - PMC - PubMed
-
- Schuh R, Aicher W, Gaul U, Cote S, Preiss A, Maier D, Seifert E, Nauber U, Schroder C, Kemler R. A conserved family of nuclear proteins containing structural elements of the finger protein encoded by Kruppel, a Drosophila segmentation gene. Cell. 1986;47:1025–1032. doi: 10.1016/0092-8674(86)90817-2. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

