A humanized TCR retaining authentic specificity and affinity conferred potent anti-tumour cytotoxicity
- PMID: 29645087
- PMCID: PMC6099166
- DOI: 10.1111/imm.12935
A humanized TCR retaining authentic specificity and affinity conferred potent anti-tumour cytotoxicity
Abstract
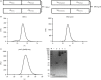
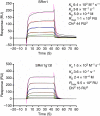
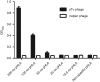
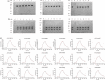
The affinity of T-cell receptor (TCR) determines the efficacy of TCR-based immunotherapy. By using human leucocyte antigen (HLA)-A*02 transgenic mice, a TCR was generated previously specific for human tumour testis antigen peptide MAGE-A3112-120 (KVAELVHFL) HLA-A*02 complex. We developed an approach to humanize the murine TCR by replacing the mouse framework with sequences of folding optimized human TCR variable domains for retaining binding affinity. The resultant humanized TCR exhibited higher affinity and conferred better anti-tumour activity than its parent murine MAGE-A3 TCR (SRm1). In addition, the affinity of humanized TCR was enhanced further to achieve improved T-cell activation. Our studies demonstrated that the human TCR variable domain frameworks could provide support for complementarity-determining regions from a murine TCR, and retain the original binding activity. It could be used as a generic approach of TCR humanization.
Keywords: T-cell; activation; humanized TCR; immunogenicity; murine TCR.
© 2018 John Wiley & Sons Ltd.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

