Identifying New Small Proteins in Escherichia coli
- PMID: 29645342
- PMCID: PMC6001520
- DOI: 10.1002/pmic.201700064
Identifying New Small Proteins in Escherichia coli
Abstract
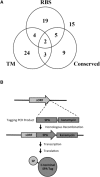
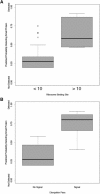
The number of small proteins (SPs) encoded in the Escherichia coli genome is unknown, as current bioinformatics and biochemical techniques make short gene and small protein identification challenging. One method of small protein identification involves adding an epitope tag to the 3' end of a short open reading frame (sORF) on the chromosome, with synthesis confirmed by immunoblot assays. In this study, this strategy was used to identify new E. coli small proteins, tagging 80 sORFs in the E. coli genome, and assayed for protein synthesis. The selected sORFs represent diverse sequence characteristics, including degrees of sORF conservation, predicted transmembrane domains, sORF direction with respect to flanking genes, ribosome binding site (RBS) prediction, and ribosome profiling results. Of 80 sORFs, 36 resulted in encoded synthesized proteins-a 45% success rate. Modeling of detected versus non-detected small proteins analysis showed predictions based on RBS prediction, transcription data, and ribosome profiling had statistically-significant correlation with protein synthesis; however, there was no correlation between current sORF annotation and protein synthesis. These results suggest substantial numbers of small proteins remain undiscovered in E. coli, and existing bioinformatics techniques must continue to improve to facilitate identification.
Keywords: SPA-tagging; small proteins.
© 2018 Towson University. Proteomics Published by WILEY-VCH Verlag GmbH & Co. KGaA, Weinheim.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

