Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning
- PMID: 29648610
- PMCID: PMC5946831
- DOI: 10.1093/gigascience/giy037
Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning
Erratum in
-
Correction to: Chiron: translating nanopore raw signal directly into nucleotide sequence using deep learning.Gigascience. 2019 May 1;8(5):giz049. doi: 10.1093/gigascience/giz049. Gigascience. 2019. PMID: 31077312 Free PMC article. No abstract available.
Abstract
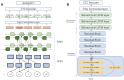
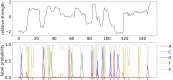
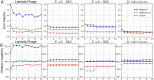
Sequencing by translocating DNA fragments through an array of nanopores is a rapidly maturing technology that offers faster and cheaper sequencing than other approaches. However, accurately deciphering the DNA sequence from the noisy and complex electrical signal is challenging. Here, we report Chiron, the first deep learning model to achieve end-to-end basecalling and directly translate the raw signal to DNA sequence without the error-prone segmentation step. Trained with only a small set of 4,000 reads, we show that our model provides state-of-the-art basecalling accuracy, even on previously unseen species. Chiron achieves basecalling speeds of more than 2,000 bases per second using desktop computer graphics processing units.
Figures
References
-
- Ashton PM, Nair S, Dallman T, et al.. MinION nanopore sequencing identifies the position and structure of a bacterial antibiotic resistance island. Nature Biotechnology. 2014;33(3):296–300. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

