Defining Driver DNA Methylation Changes in Human Cancer
- PMID: 29649096
- PMCID: PMC5979276
- DOI: 10.3390/ijms19041166
Defining Driver DNA Methylation Changes in Human Cancer
Abstract
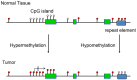
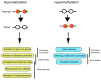
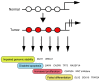
Human malignant tumors are characterized by pervasive changes in the patterns of DNA methylation. These changes include a globally hypomethylated tumor cell genome and the focal hypermethylation of numerous 5'-cytosine-phosphate-guanine-3' (CpG) islands, many of them associated with gene promoters. It has been challenging to link specific DNA methylation changes with tumorigenesis in a cause-and-effect relationship. Some evidence suggests that cancer-associated DNA hypomethylation may increase genomic instability. Promoter hypermethylation events can lead to silencing of genes functioning in pathways reflecting hallmarks of cancer, including DNA repair, cell cycle regulation, promotion of apoptosis or control of key tumor-relevant signaling networks. A convincing argument for a tumor-driving role of DNA methylation can be made when the same genes are also frequently mutated in cancer. Many of the most commonly hypermethylated genes encode developmental transcription factors, the methylation of which may lead to permanent gene silencing. Inactivation of such genes will deprive the cells in which the tumor may initiate from the option of undergoing or maintaining lineage differentiation and will lock them into a perpetuated stem cell-like state thus providing an additional window for cell transformation.
Keywords: 5-methylcytosine; DNA methylation; cell differentiation; genomic instability; hallmarks of cancer.
Conflict of interest statement
The author declares no conflict of interest.
Figures
References
-
- Ehrlich M., Lacey M. DNA hypomethylation and hemimethylation in cancer. Adv. Exp. Med. Biol. 2013;754:31–56. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

