Stable centrosomal roots disentangle to allow interphase centriole independence
- PMID: 29649211
- PMCID: PMC5918241
- DOI: 10.1371/journal.pbio.2003998
Stable centrosomal roots disentangle to allow interphase centriole independence
Update in
-
cNap1 bridges centriole contact sites to maintain centrosome cohesion.PLoS Biol. 2022 Oct 25;20(10):e3001854. doi: 10.1371/journal.pbio.3001854. eCollection 2022 Oct. PLoS Biol. 2022. PMID: 36282799 Free PMC article.
Abstract
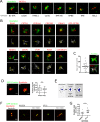
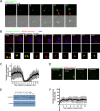
The centrosome is a non-membrane-bound cellular compartment consisting of 2 centrioles surrounded by a protein coat termed the pericentriolar material (PCM). Centrioles generally remain physically associated together (a phenomenon called centrosome cohesion), yet how this occurs in the absence of a bounding lipid membrane is unclear. One model posits that pericentriolar fibres formed from rootletin protein directly link centrioles, yet little is known about the structure, biophysical properties, or assembly kinetics of such fibres. Here, I combine live-cell imaging of endogenously tagged rootletin with cell fusion and find previously unrecognised plasticity in centrosome cohesion. Rootletin forms large, diffusionally stable bifurcating fibres, which amass slowly on mature centrioles over many hours from anaphase. Nascent centrioles (procentrioles), in contrast, do not form roots and must be licensed to do so through polo-like kinase 1 (PLK1) activity. Transient separation of roots accompanies centriolar repositioning during the interphase, suggesting that centrioles organize as independent units, each containing discrete roots. Indeed, forced induction of duplicate centriole pairs allows independent reshuffling of individual centrioles between the pairs. Therefore collectively, these findings suggest that progressively nucleated polymers mediate the dynamic association of centrioles as either 1 or 2 interphase centrosomes, with implications for the understanding of how non-membrane-bound organelles self-organise.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Schmidt KN, Kuhns S, Neuner A, Hub B, Zentgraf H, Pereira G. Cep164 mediates vesicular docking to the mother centriole during early steps of ciliogenesis. J Cell Biol. 2012;199: 1083–1101. doi: 10.1083/jcb.201202126 - DOI - PMC - PubMed
-
- Bahe S, Stierhof Y-D, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J Cell Biol. 2005;171: 27–33. doi: 10.1083/jcb.200504107 - DOI - PMC - PubMed
-
- Bernhard W, De Harven E. [Electron microscopic study of the ultrastructure of centrioles in vertebra]. Z Zellforsch Mikrosk Anat Vienna Austria 1948. 1956;45: 378–398. - PubMed
-
- Bornens M, Paintrand M, Berges J, Marty MC, Karsenti E. Structural and chemical characterization of isolated centrosomes. Cell Motil Cytoskeleton. 1987;8: 238–249. doi: 10.1002/cm.970080305 - DOI - PubMed
-
- Graser S, Stierhof Y-D, Nigg EA. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J Cell Sci. 2007;120: 4321–4331. doi: 10.1242/jcs.020248 - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

