New insights into the transposition mechanisms of IS6110 and its dynamic distribution between Mycobacterium tuberculosis Complex lineages
- PMID: 29649213
- PMCID: PMC5896891
- DOI: 10.1371/journal.pgen.1007282
New insights into the transposition mechanisms of IS6110 and its dynamic distribution between Mycobacterium tuberculosis Complex lineages
Abstract
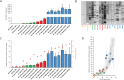
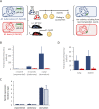
The insertion Sequence IS6110, only present in the pathogens of the Mycobacterium tuberculosis Complex (MTBC), has been the gold-standard epidemiological marker for TB for more than 25 years, but biological implications of IS6110 transposition during MTBC adaptation to humans remain elusive. By studying 2,236 clinical isolates typed by IS6110-RFLP and covering the MTBC, we remarked a lineage-specific content of IS6110 being higher in modern globally distributed strains. Once observed the IS6110 distribution in the MTBC, we selected representative isolates and found a correlation between the normalized expression of IS6110 and its abundance in MTBC chromosomes. We also studied the molecular regulation of IS6110 transposition and we found a synergistic action of two post-transcriptional mechanisms: a -1 ribosomal frameshift and a RNA pseudoknot which interferes translation. The construction of a transcriptionally active transposase resulted in 20-fold increase of the transposition frequency. Finally, we examined transposition in M. bovis and M. tuberculosis during laboratory starvation and in a mouse infection model of TB. Our results shown a higher transposition in M. tuberculosis, that preferably happens during TB infection in mice and after one year of laboratory culture, suggesting that IS6110 transposition is dynamically adapted to the host and to adverse growth conditions.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Paulson T. Epidemiology: A mortal foe. Nature. 2013;502(7470):S2–3. doi: 10.1038/502S2a . - DOI - PubMed
-
- WHO. Global Tuberculosis Report 2017. 2017.
-
- Gonzalo-Asensio J, Aguilo N, Marinova D, Martin C. Breaking Transmission with Vaccines: The Case of Tuberculosis. Microbiol Spectr. 2017;5(4). doi: 10.1128/microbiolspec.MTBP-0001-2016 . - DOI - PMC - PubMed
-
- Hedges RW, Jacob AE. Transposition of ampicillin resistance from RP4 to other replicons. Mol Gen Genet. 1974;132(1):31–40. . - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

