HPV DNA methylation at the early promoter and E1/E2 integrity: A comparison between HPV16, HPV18 and HPV45 in cervical cancer
- PMID: 29649654
- PMCID: PMC6046686
- DOI: 10.1016/j.pvr.2018.04.002
HPV DNA methylation at the early promoter and E1/E2 integrity: A comparison between HPV16, HPV18 and HPV45 in cervical cancer
Abstract
Objectives: To compare and describe type-specific characteristics of HPV16, HPV18 and HPV45 in cervical cancer with respect to 3'LCR methylation and disruption of E1/E2.
Methods: The methylation level of 137 cervical cancer samples (70 with HPV16, 37 with HPV18, and 30 with HPV45) of Brazilian patients was analyzed by pyrosequencing. PCR amplifications were performed to characterize E1 and E2 disruption as an episomal surrogate.
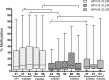
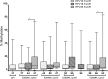
Results: The 3'LCR of HPV16 showed a higher methylation at all CpG sites (7%, 9%, 11%, 10% and 10%) than homologous HPV18 regions (4%, 5%. 6%, 9% and 5%) and HPV45 regions (7%, 7% and 5%). Presence of intact E1/E2 was associated with higher HPV16 and HPV18 methylation levels at all CpG sites (p < 0.05). Disruption of E1/E2 was more frequently found in HPV45 (97%) and HPV18 (84%) than in HPV16 DNA (30%). HPV16 disruption was more frequently found in E1 (48%) unlike HPV18, where it was found in E2 (61%). Concomitant disruption of E1/E2 was most frequent in HPV45 (72%).
Conclusions: The findings showed a higher methylation associated with intact E1/E2 for HPV16 and HPV18. The closely phylogenetic related HPV18 and HPV45 share a similar methylation level and the frequency of viral genome disruption.
Keywords: HPV genome; Human papillomavirus; Invasive cervical cancer; Methylation; Pyrosequencing; Viral genome integration.
Copyright © 2018 The Authors. Published by Elsevier B.V. All rights reserved.
Figures



References
-
- Doorbar J. Molecular biology of human papillomavirus infection and cervical cancer. Clin. Sci. 2006;110:525–541. - PubMed
-
- Mark O′Connor S.-Y.C., Bernard Hans-Ulrich. Transcription Factor Binding Sites in the Long Control Region of Genital HPVs. In: Myers G., Bernard H.U., Delius H., Baker K., Icenogle J., Halpern A., Wheeler C., editors. Human Papillomaviruses, 1995 Compendium, Part III. Los Alamos National Laboratory; Los Alamos, NMex: 1995. pp. 21–40.
-
- Bernard H.U. Gene expression of genital human papillomaviruses and considerations on potential antiviral approaches. Antivir. Ther. 2002;7:219–237. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

