Defective protein repair under methionine sulfoxide A deletion drives autophagy and ARE-dependent gene transcription
- PMID: 29649787
- PMCID: PMC5953240
- DOI: 10.1016/j.redox.2018.04.001
Defective protein repair under methionine sulfoxide A deletion drives autophagy and ARE-dependent gene transcription
Abstract
Objective: Reduction of oxidized methionines is emerging as a major protein repair pathway. The lack of methionine sulfoxide reductase A (MsrA) exacerbates cardiovascular disease phenotypes driven by increased oxidative stress. However, the role of MsrA on maintaining cellular homeostasis in the absence of excessive oxidative stress is less well understood.
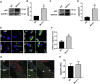
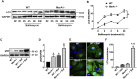
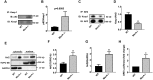
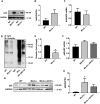
Methods and results: Constitutive genetic deletion of MsrA increased formation of p62-containing protein aggregates, activated autophagy, and decreased a marker of apoptosis in vascular smooth muscle cells (VSMC). The association of Keap1 with p62 was augmented in MsrA-/- VSMC. Keap1 targets the transcription factor Nrf2, which regulates antioxidant genes, for proteasomal degradation. However, in MsrA-/- VSMC, the association of Nrf2 with Keap1 was diminished. Whereas Nrf2 mRNA levels were not decreased in MsrA-/- VSMC, we detected decreased ubiquitination of Nrf2 and a corresponding increase in total Nrf2 protein in the absence of biochemical markers of oxidative stress. Moreover, nuclear-localized Nrf2 was increased under MsrA deficiency, resulting in upregulation of Nrf2-dependent transcriptional activity. Consequently, transcription, protein levels and enzymatic activity of glutamate-cysteine ligase and glutathione reductase were greatly augmented in MsrA-/- VSMC.
Summary: Our findings demonstrate that reversal of methionine oxidation is required for maintenance of cellular homeostasis in the absence of increased oxidative stress. These data provide the first link between autophagy and activation of Nrf2 in the setting of MsrA deletion.
Keywords: Autophagy; Methionine; Methionine sulfoxide reductase; Nrf2; Smooth muscle; Ubiquitination.
Published by Elsevier B.V.
Figures



Similar articles
-
l-Methionine activates Nrf2-ARE pathway to induce endogenous antioxidant activity for depressing ROS-derived oxidative stress in growing rats.J Sci Food Agric. 2019 Aug 15;99(10):4849-4862. doi: 10.1002/jsfa.9757. Epub 2019 May 13. J Sci Food Agric. 2019. PMID: 31001831
-
Methionine sulfoxide reductase A attenuates heme oxygenase-1 induction through inhibition of Nrf2 activation.Arch Biochem Biophys. 2012 Dec 15;528(2):134-40. doi: 10.1016/j.abb.2012.09.012. Epub 2012 Oct 2. Arch Biochem Biophys. 2012. PMID: 23036869
-
Methionine Sulfoxide Reductase A (MsrA) and Its Function in Ubiquitin-Like Protein Modification in Archaea.mBio. 2017 Sep 5;8(5):e01169-17. doi: 10.1128/mBio.01169-17. mBio. 2017. PMID: 28874471 Free PMC article.
-
Novel target for treating Alzheimer's Diseases: Crosstalk between the Nrf2 pathway and autophagy.Ageing Res Rev. 2021 Jan;65:101207. doi: 10.1016/j.arr.2020.101207. Epub 2020 Nov 1. Ageing Res Rev. 2021. PMID: 33144123 Review.
-
Nrf2-Keap1 signaling in oxidative and reductive stress.Biochim Biophys Acta Mol Cell Res. 2018 May;1865(5):721-733. doi: 10.1016/j.bbamcr.2018.02.010. Epub 2018 Feb 27. Biochim Biophys Acta Mol Cell Res. 2018. PMID: 29499228 Review.
Cited by
-
Functional Amino Acids and Autophagy: Diverse Signal Transduction and Application.Int J Mol Sci. 2021 Oct 22;22(21):11427. doi: 10.3390/ijms222111427. Int J Mol Sci. 2021. PMID: 34768858 Free PMC article. Review.
-
Acacetin exerts antioxidant potential against atherosclerosis through Nrf2 pathway in apoE-/- Mice.J Cell Mol Med. 2021 Jan;25(1):521-534. doi: 10.1111/jcmm.16106. Epub 2020 Nov 26. J Cell Mol Med. 2021. PMID: 33241629 Free PMC article.
-
The transcription factor Sox7 modulates endocardiac cushion formation contributed to atrioventricular septal defect through Wnt4/Bmp2 signaling.Cell Death Dis. 2021 Apr 12;12(4):393. doi: 10.1038/s41419-021-03658-z. Cell Death Dis. 2021. PMID: 33846290 Free PMC article.
-
Loss of methionine sulfoxide reductases increases resistance to oxidative stress.Free Radic Biol Med. 2019 Dec;145:374-384. doi: 10.1016/j.freeradbiomed.2019.10.006. Epub 2019 Oct 10. Free Radic Biol Med. 2019. PMID: 31606431 Free PMC article.
-
Sexually dimorphic effects of methionine sulfoxide reductase A (MsrA) on murine longevity and health span during methionine restriction.Geroscience. 2023 Oct;45(5):3003-3017. doi: 10.1007/s11357-023-00857-8. Epub 2023 Jun 30. Geroscience. 2023. PMID: 37391679 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

