Enduring epigenetic landmarks define the cancer microenvironment
- PMID: 29650553
- PMCID: PMC5932604
- DOI: 10.1101/gr.229070.117
Enduring epigenetic landmarks define the cancer microenvironment
Abstract
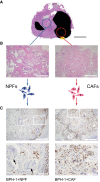
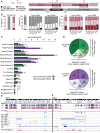
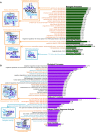
The growth and progression of solid tumors involves dynamic cross-talk between cancer epithelium and the surrounding microenvironment. To date, molecular profiling has largely been restricted to the epithelial component of tumors; therefore, features underpinning the persistent protumorigenic phenotype of the tumor microenvironment are unknown. Using whole-genome bisulfite sequencing, we show for the first time that cancer-associated fibroblasts (CAFs) from localized prostate cancer display remarkably distinct and enduring genome-wide changes in DNA methylation, significantly at enhancers and promoters, compared to nonmalignant prostate fibroblasts (NPFs). Differentially methylated regions associated with changes in gene expression have cancer-related functions and accurately distinguish CAFs from NPFs. Remarkably, a subset of changes is shared with prostate cancer epithelial cells, revealing the new concept of tumor-specific epigenome modifications in the tumor and its microenvironment. The distinct methylome of CAFs provides a novel epigenetic hallmark of the cancer microenvironment and promises new biomarkers to improve interpretation of diagnostic samples.
© 2018 Pidsley et al.; Published by Cold Spring Harbor Laboratory Press.
Figures





References
-
- Albino D, Longoni N, Curti L, Mello-Grand M, Pinton S, Civenni G, Thalmann G, D'Ambrosio G, Sarti M, Sessa F, et al. 2012. ESE3/EHF controls epithelial cell differentiation and its loss leads to prostate tumors with mesenchymal and stem-like features. Cancer Res 72: 2889–2900. - PubMed
-
- Ao M, Franco OE, Park D, Raman D, Williams K, Hayward SW. 2007. Cross-talk between paracrine-acting cytokine and chemokine pathways promotes malignancy in benign human prostatic epithelium. Cancer Res 67: 4244–4253. - PubMed
-
- Ashida S, Orloff MS, Bebek G, Zhang L, Zheng P, Peehl DM, Eng C. 2012. Integrated analysis reveals critical genomic regions in prostate tumor microenvironment associated with clinicopathologic phenotypes. Clin Cancer Res 18: 1578–1587. - PubMed
-
- Ayala G, Tuxhorn JA, Wheeler TM, Frolov A, Scardino PT, Ohori M, Wheeler M, Spitler J, Rowley DR. 2003. Reactive stroma as a predictor of biochemical-free recurrence in prostate cancer. Clin Cancer Res 9: 4792–4801. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
