Setd2 regulates quiescence and differentiation of adult hematopoietic stem cells by restricting RNA polymerase II elongation
- PMID: 29650642
- PMCID: PMC6029524
- DOI: 10.3324/haematol.2018.187708
Setd2 regulates quiescence and differentiation of adult hematopoietic stem cells by restricting RNA polymerase II elongation
Abstract
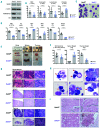
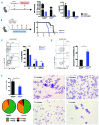
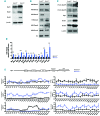
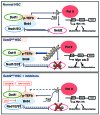
SET domain containing 2 (Setd2), encoding a histone methyltransferase, is associated with many hematopoietic diseases when mutated. By generating a novel exon 6 conditional knockout mouse model, we describe an essential role of Setd2 in maintaining the adult hematopoietic stem cells. Loss of Setd2 results in leukopenia, anemia, and increased platelets accompanied by hypocellularity, erythroid dysplasia, and mild fibrosis in bone marrow. Setd2 knockout mice show significantly decreased hematopoietic stem and progenitor cells except for erythroid progenitors. Setd2 knockout hematopoietic stem cells fail to establish long-term bone marrow reconstitution after transplantation because of the loss of quiescence, increased apoptosis, and reduced multiple-lineage terminal differentiation potential. Bioinformatic analysis revealed that the hematopoietic stem cells exit from quiescence and commit to differentiation, which lead to hematopoietic stem cell exhaustion. Mechanistically, we attribute an important Setd2 function in murine adult hematopoietic stem cells to the inhibition of the Nsd1/2/3 transcriptional complex, which recruits super elongation complex and controls RNA polymerase II elongation on a subset of target genes, including Myc Our results reveal a critical role of Setd2 in regulating quiescence and differentiation of hematopoietic stem cells through restricting the NSDs/SEC mediated RNA polymerase II elongation.
Copyright© 2018 Ferrata Storti Foundation.
Figures




Comment in
-
Linking histone methylation, transcription rates, and stem cell robustness.Haematologica. 2018 Jul;103(7):1093. doi: 10.3324/haematol.2018.196089. Haematologica. 2018. PMID: 29970491 Free PMC article. No abstract available.
References
-
- Kiel MJ, Yilmaz OH, Iwashita T, et al. SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell. 2005;121(7):1109–1121. - PubMed
-
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

