HEx: A heterologous expression platform for the discovery of fungal natural products
- PMID: 29651464
- PMCID: PMC5895447
- DOI: 10.1126/sciadv.aar5459
HEx: A heterologous expression platform for the discovery of fungal natural products
Abstract
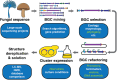
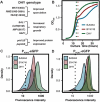
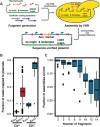
For decades, fungi have been a source of U.S. Food and Drug Administration-approved natural products such as penicillin, cyclosporine, and the statins. Recent breakthroughs in DNA sequencing suggest that millions of fungal species exist on Earth, with each genome encoding pathways capable of generating as many as dozens of natural products. However, the majority of encoded molecules are difficult or impossible to access because the organisms are uncultivable or the genes are transcriptionally silent. To overcome this bottleneck in natural product discovery, we developed the HEx (Heterologous EXpression) synthetic biology platform for rapid, scalable expression of fungal biosynthetic genes and their encoded metabolites in Saccharomyces cerevisiae. We applied this platform to 41 fungal biosynthetic gene clusters from diverse fungal species from around the world, 22 of which produced detectable compounds. These included novel compounds with unexpected biosynthetic origins, particularly from poorly studied species. This result establishes the HEx platform for rapid discovery of natural products from any fungal species, even those that are uncultivable, and opens the door to discovery of the next generation of natural products.
Figures


References
-
- Newman D. J., Cragg G. M., Natural products as sources of new drugs from 1981 to 2014. J. Nat. Prod. 79, 629–661 (2016). - PubMed
-
- Schueffler A., Anke T., Fungal natural products in research and development. Nat. Prod. Rep. 31, 1425–1448 (2014). - PubMed
-
- Wiemann P., Keller N. P., Strategies for mining fungal natural products. J. Ind. Microbiol. Biotechnol. 41, 301–313 (2014). - PubMed
-
- Blackwell M., The fungi: 1, 2, 3 … 5.1 million species? Am. J. Bot. 98, 426–438 (2011). - PubMed
-
- Inglis D. O., Binkley J., Skrzypek M. S., Arnaud M. B., Cerqueira G. C., Shah P., Wymore F., Wortman J. R., Sherlock G., Comprehensive annotation of secondary metabolite biosynthetic genes and gene clusters of Aspergillus nidulans, A. fumigatus, A. niger and A. oryzae. BMC Microbiol. 13, 91 (2013). - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

