In the Telencephalon, GluN2C NMDA Receptor Subunit mRNA is Predominately Expressed in Glial Cells and GluN2D mRNA in Interneurons
- PMID: 29651654
- PMCID: PMC6349034
- DOI: 10.1007/s11064-018-2526-7
In the Telencephalon, GluN2C NMDA Receptor Subunit mRNA is Predominately Expressed in Glial Cells and GluN2D mRNA in Interneurons
Abstract
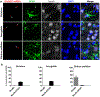
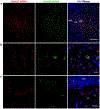
N-methyl-D-aspartate receptors (NMDARs) are widely distributed in the brain with high concentrations in the telencephalon where they modulate synaptic plasticity, working memory, and other functions. While the actions of the predominate GluN2 NMDAR subunits, GluN2A and GluN2B are relatively well understood, the function of GluN2C and GluN2D subunits in the telencephalon is largely unknown. To better understand the possible role of GluN2C subunits, we used fluorescence in situ hybridization (FISH) together with multiple cell markers to define the distribution and type of cells expressing GluN2C mRNA. Using a GluN2C-KO mouse as a negative control, GluN2C mRNA expression was only found in non-neuronal cells (NeuN-negative cells) in the hippocampus, striatum, amygdala, and cerebral cortex. For these regions, a significant fraction of GFAP-positive cells also expressed GluN2C mRNA. Overall, for the telencephalon, the globus pallidus and olfactory bulb were the only regions where GluN2C was expressed in neurons. In contrast to GluN2C, GluN2D subunit mRNA colocalized with neuronal and not astrocyte markers or GluN2C mRNA in the telencephalon (except for the globus pallidus). GluN2C mRNA did, however, colocalize with GluN2D in the thalamus where neuronal GluN2C expression is found. These findings strongly suggest that GluN2C has a very distinct function in the telencephalon compared to its role in other brain regions and compared to other GluN2-containing NMDARs. NMDARs containing GluN2C may have a specific role in regulating L-glutamate or D-serine release from astrocytes in response to L-glutamate spillover from synaptic activity.
Keywords: Astrocyte; Cortex; GluN2C; GluN2D; Hippocampus; L-glutamate; N-methyl-D-aspartate receptor; mRNA.
Figures








References
-
- Monyer H, Burnashev N, Laurie DJ, Sakmann B and Seeburg PH, (1994) Developmental and regional expression in the rat brain and functional properties of four NMDA receptors. Neuron. 12, 529–540. - PubMed
-
- Mishina M, Mori H, Araki K, Kushiya E, Meguro H, Kutsuwada T, Kashiwabuchi N, Ikeda K, Nagasawa M and Yamazaki M, (1993) Molecular and functional diversity of the NMDA receptor channel. Ann. N. Y. Acad. Sci 707, 136–152. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

