Microtubule assembly governed by tubulin allosteric gain in flexibility and lattice induced fit
- PMID: 29652248
- PMCID: PMC5945277
- DOI: 10.7554/eLife.34353
Microtubule assembly governed by tubulin allosteric gain in flexibility and lattice induced fit
Abstract
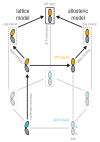
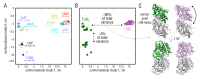
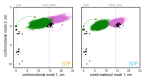
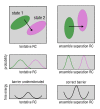
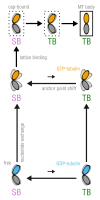
Microtubules (MTs) are key components of the cytoskeleton and play a central role in cell division and development. MT assembly is known to be associated with a structural change in [Formula: see text]-tubulin dimers from kinked to straight conformations. How GTP binding renders individual dimers polymerization-competent, however, is still unclear. Here, we have characterized the conformational dynamics and energetics of unassembled tubulin using atomistic molecular dynamics and free energy calculations. Contrary to existing allosteric and lattice models, we find that GTP-tubulin favors a broad range of almost isoenergetic curvatures, whereas GDP-tubulin has a much lower bending flexibility. Moreover, irrespective of the bound nucleotide and curvature, two conformational states exist differing in location of the anchor point connecting the monomers that affects tubulin bending, with one state being strongly favored in solution. Our findings suggest a new combined model in which MTs incorporate and stabilize flexible GTP-dimers with a specific anchor point state.
Keywords: dynamic instability; microtubule; molecular biophysics; molecular dynamics; none; structural biology; tubulin.
© 2018, Igaev et al.
Conflict of interest statement
MI, HG No competing interests declared
Figures



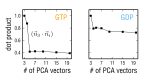


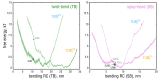
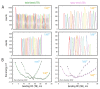
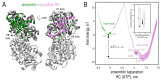

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

