RhoGAPp190: A potential player in tbph-mediated neurodegeneration in Drosophila
- PMID: 29652933
- PMCID: PMC5898758
- DOI: 10.1371/journal.pone.0195845
RhoGAPp190: A potential player in tbph-mediated neurodegeneration in Drosophila
Abstract
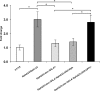
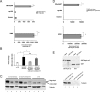
TDP-43 is an ubiquitous and highly conserved ribonucleoprotein involved in several cellular processes including pre-mRNA splicing, transcription, mRNA stability and transport. Notwithstanding the evidence of TDP-43 involvement in the pathogenesis of different neurodegenerative disorders (i.e. ALS and FTLD), the underlying mechanisms are still unclear. Given the high degree of functional similarity between the human and fly orthologs of TDP-43, Drosophila melanogaster is a simple and useful model to study the pathophysiological role of this protein in vivo. It has been demonstrated that the depletion of the TDP-43 fly ortholog (tbph) induces deficient locomotive behaviors and reduces life span and anatomical defects at the neuromuscular junction. In this study, using the known binding specificity of TDP-43/tbph for (UG) repeated sequences, we performed a bioinformatic screening for fly genes with at least 6 (TG) repeats in a row within the 3'-UTR regions in order to identify the genes that might be regulated by this factor. Among these genes, we were able to identify RhoGAPp190 as a potential target of the tbph-mediated neurodegeneration. RhoGAPp190 is a negative regulator of Drosophila RhoA, a GTPase protein implicated in the fine modulation of critical cellular processes including axon branch stability and motor axon defasciculation at muscle level and cognitive processes. We were able to demonstrate that the RhoGAPp190 expression is upregulated in a tbph-null fly model, providing evidence that this deregulation is associated to tbph silencing. Our results introduce RhoGAPp190 as a novel potential mediator in the complex scenario of events resulting from in vivo tbph loss-of-function.
Conflict of interest statement
Figures



References
-
- Ratti A, Buratti E. Physiological functions and pathobiology of TDP-43 and FUS/TLS proteins. J Neurochem. 2016;138 Suppl 1:95–111. doi: 10.1111/jnc.13625 . - DOI - PubMed
-
- Arai T, Hasegawa M, Akiyama H, Ikeda K, Nonaka T, Mori H, et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem Biophys Res Commun. 2006;351(3):602–11. Epub 2006/11/07. S0006-291X(06)02318-7 [pii] doi: 10.1016/j.bbrc.2006.10.093 . - DOI - PubMed
-
- Neumann M, Sampathu DM, Kwong LK, Truax AC, Micsenyi MC, Chou TT, et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314(5796):130–3. doi: 10.1126/science.1134108 . - DOI - PubMed
-
- Ling SC, Polymenidou M, Cleveland DW. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79(3):416–38. doi: 10.1016/j.neuron.2013.07.033 ; PubMed Central PMCID: PMC4411085. - DOI - PMC - PubMed
-
- Van Langenhove T, van der Zee J, Van Broeckhoven C. The molecular basis of the frontotemporal lobar degeneration-amyotrophic lateral sclerosis spectrum. Annals of medicine. 2012;44(8):817–28. doi: 10.3109/07853890.2012.665471 ; PubMed Central PMCID: PMC3529157. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

