Amyloid assembly and disassembly
- PMID: 29654159
- PMCID: PMC5963839
- DOI: 10.1242/jcs.189928
Amyloid assembly and disassembly
Abstract
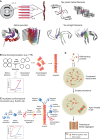
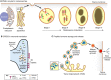
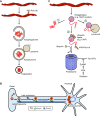
Amyloid fibrils are protein homopolymers that adopt diverse cross-β conformations. Some amyloid fibrils are associated with the pathogenesis of devastating neurodegenerative disorders, including Alzheimer's disease and Parkinson's disease. Conversely, functional amyloids play beneficial roles in melanosome biogenesis, long-term memory formation and release of peptide hormones. Here, we showcase advances in our understanding of amyloid assembly and structure, and how distinct amyloid strains formed by the same protein can cause distinct neurodegenerative diseases. We discuss how mutant steric zippers promote deleterious amyloidogenesis and aberrant liquid-to-gel phase transitions. We also highlight effective strategies to combat amyloidogenesis and related toxicity, including: (1) small-molecule drugs (e.g. tafamidis) to inhibit amyloid formation or (2) stimulate amyloid degradation by the proteasome and autophagy, and (3) protein disaggregases that disassemble toxic amyloid and soluble oligomers. We anticipate that these advances will inspire therapeutics for several fatal neurodegenerative diseases.
Keywords: Amyloid; Autophagy; Disaggregase; Neurodegeneration; Prion.
© 2018. Published by The Company of Biologists Ltd.
Conflict of interest statement
Competing interestsThe authors declare no competing or financial interests.
Figures

References
-
- Abedini A., Plesner A., Cao P., Ridgway Z., Zhang J., Tu L.-H., Middleton C. T., Chao B., Sartori D. J., Meng F. et al. (2016). Time-resolved studies define the nature of toxic IAPP intermediates, providing insight for anti-amyloidosis therapeutics. 5, e12977 10.7554/eLife.12977 - DOI - PMC - PubMed
-
- Abisambra J., Jinwal U. K., Miyata Y., Rogers J., Blair L., Li X., Seguin S. P., Wang L., Jin Y., Bacon J. et al. (2013). Allosteric heat shock protein 70 inhibitors rapidly rescue synaptic plasticity deficits by reducing aberrant tau. 74, 367-374. 10.1016/j.biopsych.2013.02.027 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

