Evidence that the metabolite repair enzyme NAD(P)HX epimerase has a moonlighting function
- PMID: 29654173
- PMCID: PMC5938422
- DOI: 10.1042/BSR20180223
Evidence that the metabolite repair enzyme NAD(P)HX epimerase has a moonlighting function
Abstract
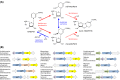
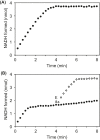
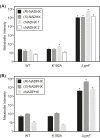
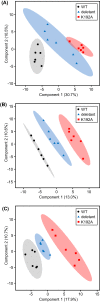
NAD(P)H-hydrate epimerase (EC 5.1.99.6) is known to help repair NAD(P)H hydrates (NAD(P)HX), which are damage products existing as R and S epimers. The S epimer is reconverted to NAD(P)H by a dehydratase; the epimerase facilitates epimer interconversion. Epimerase deficiency in humans causes a lethal disorder attributed to NADHX accumulation. However, bioinformatic evidence suggest caution about this attribution by predicting that the epimerase has a second function connected to vitamin B6 (pyridoxal 5'-phosphate and related compounds). Specifically, (i) the epimerase is fused to a B6 salvage enzyme in plants, (ii) epimerase genes cluster on the chromosome with B6-related genes in bacteria, and (iii) epimerase and B6-related genes are coexpressed in yeast and Arabidopsis The predicted second function was explored in Escherichia coli, whose epimerase and dehydratase are fused and encoded by yjeF The putative NAD(P)HX epimerase active site has a conserved lysine residue (K192 in E. coli YjeF). Changing this residue to alanine cut in vitro epimerase activity by ≥95% but did not affect dehydratase activity. Mutant cells carrying the K192A mutation had essentially normal NAD(P)HX dehydratase activity and NAD(P)HX levels, showing that the mutation had little impact on NAD(P)HX repair in vivo However, these cells showed metabolome changes, particularly in amino acids, which exceeded those in cells lacking the entire yjeF gene. The K192A mutant cells also had reduced levels of 'free' (i.e. weakly bound or unbound) pyridoxal 5'-phosphate. These results provide circumstantial evidence that the epimerase has a metabolic function beyond NAD(P)HX repair and that this function involves vitamin B6.
Keywords: AIBP; NAD(P)H hydrates; NAD(P)HX; NAXE; Vitamin B6; protein moonlighting.
© 2018 The Author(s).
Conflict of interest statement
The authors declare that there are no competing interests associated with the manuscript.
Figures


Comment in
-
Genotype to phenotype mapping still needs underpinning by research in metabolism and enzymology.Biosci Rep. 2018 Jun 21;38(3):BSR20180520. doi: 10.1042/BSR20180520. Print 2018 Jun 29. Biosci Rep. 2018. PMID: 29848765 Free PMC article.
References
-
- Oppenheimer N.J. and Kaplan N.O. (1974) Glyceraldehyde-3-phosphate dehydrogenase catalyzed hydration of the 5-6 double bond of reduced β-nicotinamide adenine dinucleotide (β-NADH). Formation of β-6-hydroxy-1,4,5,6-tetrahydronicotinamide adenine dinucleotide. Biochemistry 13, 4685–4694 10.1021/bi00720a002 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

